Seg Former 3 D: an Efficient Transformer for 3D Medical Image Segmentation
SegFormer 3D: 一种高效的3D医学图像分割Transformer
Abstract
摘要
The adoption of Vision Transformers (ViTs) based architectures represents a significant advancement in 3D Medical Image (MI) segmentation, surpassing traditional Convolutional Neural Network (CNN) models by enhancing global contextual understanding. While this paradigm shift has significantly enhanced 3D segmentation performance, state-of-the-art architectures require extremely large and complex architectures with large scale computing resources for training and deployment. Furthermore, in the context of limited datasets, often encountered in medical imaging, larger models can present hurdles in both model genera liz ation and convergence. In response to these challenges and to demonstrate that lightweight models are a valuable area of research in 3D medical imaging, we present Seg Former 3 D, a hierarchical Transformer that calculates attention across multiscale volumetric features. Additionally, Seg Former 3 D avoids complex decoders and uses an all-MLP decoder to aggregate local and global attention features to produce highly accurate segmentation masks. The proposed memory efficient Transformer preserves the performance characteristics of a significantly larger model in a compact design. Seg Former 3 D democratizes deep learning for $3D$ medical image segmentation by offering a model with $33\times$ less parameters and a $13\times$ reduction in GFLOPS compared to the current state-of-theart (SOTA). We benchmark Seg Former 3 D against the current SOTA models on three widely used datasets Synapse, BRaTs, and ACDC, achieving competitive results. Code: https://github.com/OSUPCVLab/Seg Former 3 D.git
基于视觉Transformer (ViT) 架构的采用标志着3D医学图像(MI)分割领域的重大进步,通过增强全局上下文理解能力超越了传统卷积神经网络(CNN)模型。尽管这一范式转变显著提升了3D分割性能,但最先进的架构需要极其庞大复杂的结构以及大规模计算资源进行训练和部署。此外,在医学影像常见的有限数据集场景下,更大规模的模型可能在模型泛化性和收敛性方面带来挑战。为应对这些问题并证明轻量级模型在3D医学影像中的重要研究价值,我们提出了SegFormer3D——一种在多层次体素特征上计算注意力的分层Transformer。该模型摒弃复杂解码器结构,采用全MLP解码器来聚合局部与全局注意力特征,从而生成高精度分割掩码。这种内存高效的Transformer通过紧凑设计保留了更大规模模型的性能特征。相比当前最优(SOTA)模型,SegFormer3D以参数减少33倍、GFLOPS降低13倍的显著优势,推动了3D医学图像分割的深度学习平民化。我们在Synapse、BRaTs和ACDC三个广泛使用的数据集上对SegFormer3D进行基准测试,均取得具有竞争力的结果。代码:https://github.com/OSUPCVLab/SegFormer3D.git
1. Introduction
1. 引言
The emergence of deep learning in healthcare has been transformative, offering an unprecedented capacity to learn and analyze complex medical data patterns. A fundamental task in medical image analysis is 3D volumetric image segmentation that is crucial for applications such as tumor and multi-organ localization in diagnosis and treatment. The conventional approach involves employing an encoderdecoder architecture [18, 23], where the image is first transformed into a low-dimensional representation, and then the decoder maps the representation to a voxel-wise segmentation mask. However, these architectures struggle to generate accurate segmentation masks due to their limited receptive field. Recently, Transformer-based techniques have demonstrated superior segmentation performance owing to the ViT’s ability to utilize attention layers for capturing global relationships [11, 32]. This stands in sheer contrast to CNNs which exhibits local inductive bias properties.
深度学习在医疗领域的出现具有变革性意义,它提供了学习和分析复杂医疗数据模式的空前能力。医学图像分析的一项基本任务是3D体积图像分割,这对诊断和治疗中的肿瘤及多器官定位等应用至关重要。传统方法采用编码器-解码器架构 [18, 23],先将图像转换为低维表示,再由解码器将表示映射为体素级分割掩码。然而,由于感受野有限,这些架构难以生成精确的分割掩码。最近,基于Transformer的技术展现出卓越的分割性能,这得益于ViT能够利用注意力层捕捉全局关系 [11, 32]。这与CNN表现出的局部归纳偏置特性形成鲜明对比。
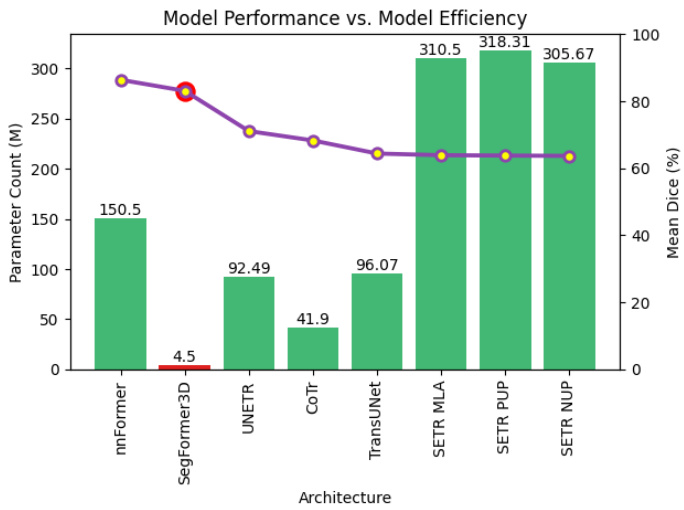
Fig. 1. Parameter Count vs. Performance on BraTs We compare Seg former 3 D to existing 3D volumetric image segmentation architectures evaluating model performance with respect to parameter count. The green bars represent model parameters while the purple plot shows the mean dice performance for each architecture. We demonstrate that at 4.5 million parameters Seg former 3 D is a highly competitive lightweight architecture for 3D medical image segmentation.
图 1: 参数量与BraTs性能对比 我们将Segformer 3D与现有3D体积图像分割架构进行对比,评估模型性能与参数量的关系。绿色柱状图表示模型参数量,紫色曲线显示各架构的平均Dice分数。实验证明,在450万参数规模下,Segformer 3D是极具竞争力的轻量级3D医学图像分割架构。
Following the seminal work of TransUnet[5] and UNETR [11], a large body of research in the medical community has been dedicated to designing Transformer based archi tec ture s that take advantage of the strong encoding capability of ViTs and the feature refinement capability of CNNs in the decoding stage. For example, [10, 11, 32] combined localized receptive field of convolutions and global attention. Despite their advantages, ViTs fail to match the genera liz ation capabilities of CNNs when trained from scratch on small-scale datasets, and because of lack of inductive bias often depends on large scale datasets for pre training [7] which are not commonly available in medical image domain. Additionally, computational efficiency of the ViTs are bounded by number of floating point operation and element wise functions in the multi head self attention block [17]. This issue is much more prominent in 3D medical imaging tasks because of the fact that the length of the converted sequence of the 3D volumetric input is considerably long. Furthermore, medical imaging data frequently exhibits repetitive structures [6], suggesting that it can be compressed, a consideration often overlooked by the 3D SOTA ViT architectures in the medical domain.
继TransUnet[5]和UNETR[11]的开创性工作之后,医学界大量研究致力于设计基于Transformer的架构,以利用ViT强大的编码能力和CNN在解码阶段的特征细化能力。例如,[10, 11, 32]结合了卷积的局部感受野和全局注意力机制。尽管具有优势,ViT在小规模数据集上从头训练时无法匹配CNN的泛化能力,并且由于缺乏归纳偏置,通常依赖于大规模预训练数据集[7],而这在医学影像领域并不常见。此外,ViT的计算效率受限于多头自注意力模块中的浮点运算数量和逐元素函数[17]。这一问题在3D医学影像任务中更为突出,因为3D体积输入转换后的序列长度相当长。更进一步,医学影像数据经常表现出重复结构[6],表明其可被压缩,而这一考量常被医学领域的3D SOTA ViT架构所忽视。
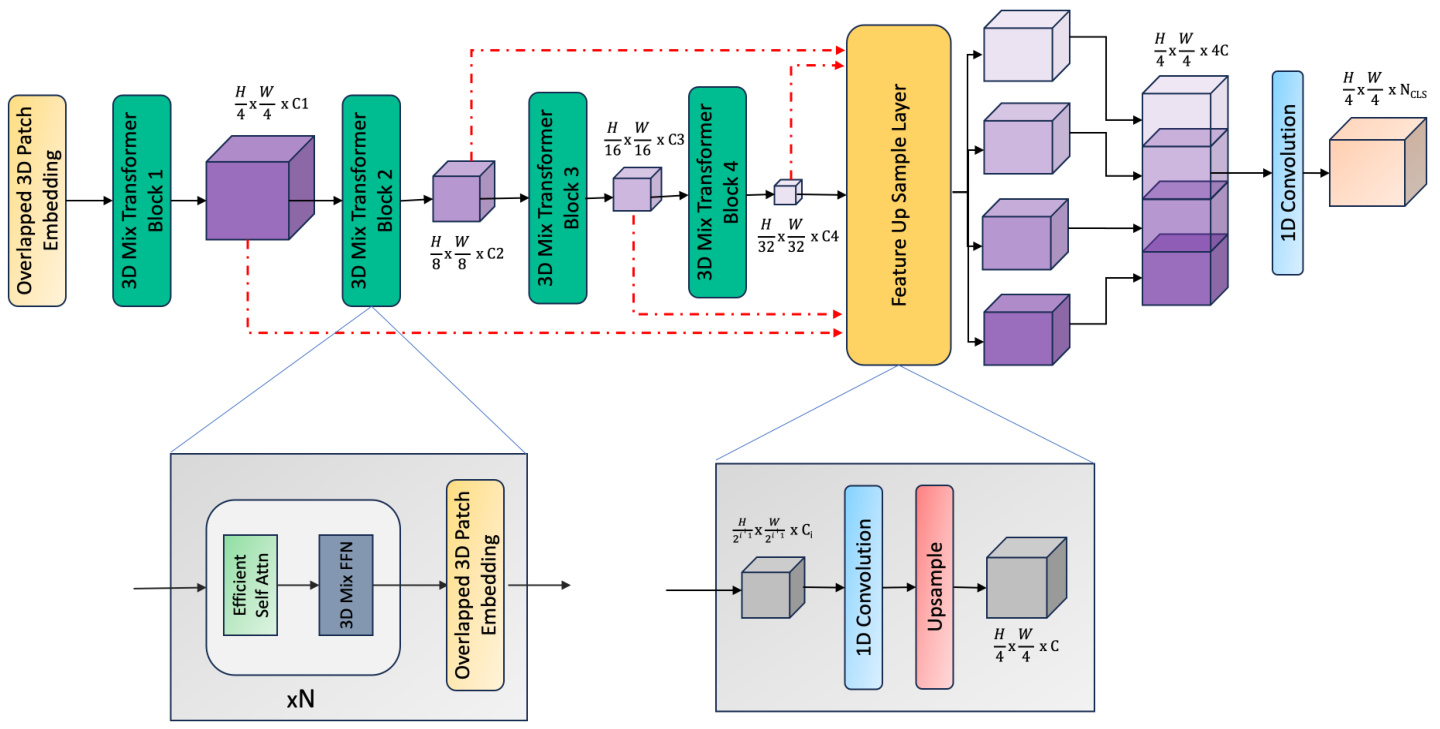
Fig. 2. Seg former 3 D Overview: The model input is a 3D volume $\mathbb{R}^{D\times C\times H\times W}$ . We extract multiscale volumetric features using a 4 stage hierarchical Transformer. An all-MLP decoder then upsamples and aggregate local and global attention features from the encoding stage to generate the final segmentation mask.
图 2: SegFormer 3D 概览:模型输入是一个 3D 体积 $\mathbb{R}^{D\times C\times H\times W}$。我们使用 4 阶段分层 Transformer 提取多尺度体积特征。随后,全 MLP 解码器对编码阶段的局部和全局注意力特征进行上采样和聚合,生成最终的分割掩码。
ally, Seg Former 3 D utilizes the overlapping patch embedding module used in [27] that preserves the local continuity of the input voxels. This embedding uses a positionalfree encoding [14] that prevents accuracy loss when there is a resolution mismatch during training and inference, which is common in medical image segmentation. To efficiently generate a high-quality segmentation mask, SegFormer3D uses an all-MLP decoder introduced in [27]. Comprehensive experiments on three benchmark datasetsSynapse[15], ACDC[1], and BRaTs[20]—validate the qualitative and quantitative effectiveness of Seg Former 3 D. Our contributions can be summarized as:
最终,SegFormer3D采用了[27]中提出的重叠块嵌入模块,该模块能保持输入体素的局部连续性。该嵌入使用了一种无位置编码[14],可避免训练与推理阶段分辨率不匹配导致的精度损失(这在医学图像分割中很常见)。为高效生成高质量分割掩膜,SegFormer3D采用了[27]提出的全MLP解码器。在三个基准数据集(Synapse[15]、ACDC[1]和BRaTs[20])上的综合实验验证了SegFormer3D在定性与定量方面的有效性。我们的贡献可归纳为:
This paper presents Seg Former 3 D, a volumetric hierarchical ViT, that extends [27] to 3D medical image segmentation tasks. Unlike vanilla ViT[7] which renders feature maps on a fixed scale, Seg former 3 D encodes feature maps at different scales of the input volume following the Pyramid Vision Transformer [26]. Our design enables the Transformer to capture a variety of coarse to fine-grained features of the input. Seg Former 3 D also utilizes an efficient self-attention module [26] that compresses the embedded sequence to a fixed ratio to significantly reduce model complexity without sacrificing performance Figure 1. Addition• We introduce a lightweight memory efficient segmentation model that preserves the performance characteristics of larger models for 3D medical imaging. • With 4.5 million parameters and 17 GFLOPS, Seg- former3D presents a $34\times$ and $13\times$ reduction in parameter count and model complexity vs SOTA. • We showcase highly competitive results without pretraining, emphasizing the generalization capabilities of lightweight ViTs and that exploring architectures like Seg former 3 D is a valuable research area in medical imaging.
本文提出SegFormer3D,一种体积分层ViT (Vision Transformer),将[27]扩展到3D医学图像分割任务。不同于固定尺度生成特征图的传统ViT[7],SegFormer3D遵循金字塔视觉Transformer[26]在输入体积的不同尺度上编码特征图。我们的设计使Transformer能捕获从粗粒度到细粒度的多样化输入特征。SegFormer3D还采用高效自注意力模块[26],将嵌入序列压缩至固定比例以显著降低模型复杂度而不牺牲性能(图1)。
• 我们提出一种轻量级内存高效的分割模型,在3D医学成像中保持大模型的性能特征
• 仅需450万参数和17 GFLOPS,SegFormer3D相比SOTA模型实现参数量34倍和模型复杂度13倍的降低
• 我们在无需预训练的情况下展示出极具竞争力的结果,凸显了轻量级ViT的泛化能力,证明探索SegFormer3D等架构是医学影像领域的重要研究方向
2. Related Work
2. 相关工作
Following the introduction of Unet[23], numerous approaches have been proposed for medical image analysis such as Dense-unet[2] and deep-supervised CNN [33]. Unet has also been extended to 3D medical image analysis, for instance, 3D-Unet[6], V-net[21], nn-Unet[13] and [8, 9, 24]. Researchers also designed hierarchical architectures to capture contextual information. In [21], Milletari et al. down sampled the volume to a lower resolution to preserve beneficial image features using V-net. Cicek et al.[6] replaced the 2D to 3D convolutions in 3D-unet. Isensee et al.[13] proposed the nn-Unet generalized segmentation architecture that can extract features at multiple scales. In [16], PGD-UNet uses deformable convolution to deal with irregular organ shapes and tumors for medical image segmentation.
继Unet[23]提出之后,医学图像分析领域涌现了大量改进方法,如Dense-unet[2]和深度监督CNN[33]。Unet还被扩展至三维医学图像分析,例如3D-Unet[6]、V-net[21]、nn-Unet[13]以及[8,9,24]等研究。学者们还设计了层次化架构来捕捉上下文信息:Milletari等人在[21]中通过V-net对体积数据进行降采样以保留有益图像特征;Cicek等人[6]将3D-Unet中的二维卷积替换为三维卷积;Isensee团队[13]提出的nn-Unet通用分割架构可实现多尺度特征提取;[16]中提出的PGD-UNet则采用可变形卷积处理医学图像分割中不规则器官形态与肿瘤。
Several recent papers have studied Transformerconvolution architectures such as TransUnet[5], Unetr[11], SwinUnetr[10], ,TransFuse[30], nnFormer[32] and LoGoNet[22]. TransUnet[5] combines Transformers and U-Net to encode image patches and decode through high resolution upsampled CNN features for localization. Hat a miz a deh et al. [11] present UNETR, a 3D model merging the long-range spatial dependencies characteristic of Transformers with the inherent CNN inductive biases in a ”U-shaped” encoder-decoder structure. In UNETR, Transformer-blocks encode features that capture consistent global representations and are subsequently integrated across various resolutions within a CNN-based decoder. LoGoNet[22] uses Large Kernel Attention (LKA) and a dual encoding strategy to capture both long-range and short-range feature dependencies for 3D medical image segmentation. Zhou et al.[32] present nnFormer, a method derived from the Swin-UNet[3] architecture. Wang et al.[25] proposed TransBTS which uses a regular convolutional encoder-decoder architecture and a Transformer layer as the bottleneck. These methods suffers from considerable model and computational complexity.
近期多篇论文研究了Transformer与卷积结合的架构,如TransUnet[5]、Unetr[11]、SwinUnetr[10]、TransFuse[30]、nnFormer[32]和LoGoNet[22]。TransUnet[5]通过Transformer编码图像块,并利用高分辨率上采样的CNN特征进行定位解码。Hatamizadeh等人[11]提出的UNETR采用三维模型,在"U型"编码器-解码器结构中融合了Transformer的长程空间依赖特性与CNN固有的归纳偏置。UNETR通过Transformer块编码获取全局一致性表征,随后在基于CNN的解码器中实现多分辨率特征融合。LoGoNet[22]采用大核注意力(LKA)和双编码策略,同时捕获长程与短程特征依赖以实现三维医学图像分割。Zhou等人[32]提出的nnFormer方法源自Swin-UNet[3]架构。Wang等人[25]开发的TransBTS采用标准卷积编码器-解码器架构,并以Transformer层作为瓶颈结构。这些方法普遍存在模型复杂度和计算量过高的问题。
3. Method
3. 方法
The adoption of Transformers has greatly improved the performance of volumetric medical image segmentation. However, current high-performing architectures prioritize overparameter iz ation for model performance, sacrificing efficiency. To demonstrate the benefits of lightweight and efficient Transformers without compromising on performance, we introduce Seg former 3 D. With 4.5 million parameters and 17 GFLOPS we show a reduction of ${\bf34}\times$ and $\pmb{13}\times$ in parameter count and complexity showcasing the significance of the proposed architecture in 3D medical image segmentation 1.
Transformer的采用极大提升了医学体图像分割的性能。然而当前高性能架构优先考虑通过过参数化提升模型表现,牺牲了效率。为证明轻量高效Transformer在不损失性能下的优势,我们提出了Segformer 3D。该模型仅需450万参数和17GFLOPS运算量,在参数量和计算复杂度上分别实现${\bf34}\times$与$\pmb{13}\times$的降低,彰显了该架构在三维医学图像分割领域的重要性[1]。
Table 1. Seg former 3 D vs SOTA in Size (M), and complexity. Seg former 3 D showcases a significant reduction in parameters and computational complexity without sacrificing on performance.
表 1: Segformer3D 与 SOTA 在参数量 (M) 和计算复杂度上的对比。Segformer3D 在保持性能的同时显著降低了参数量和计算复杂度。
| 架构 | 参数量 | GFLOPs |
|---|---|---|
| nnFormer[32] | 150.5 | 213.4 |
| TransUnet[5] | 96.07 | 88.91 |
| UNETR[11] | 92.49 | 75.76 |
| SwinUNETR[10] | 62.83 | 384.2 |
| Segformer3D (ours) | 4.51 | 17.5 |
Encoder: Using 3D medical images within the Transformer framework results in long sequence lengths which increases the computational complexity of the model. For example, a standard 3D MRI volume with dimensions of 128³ results in a sequence length of 32,768, whereas a typical 2D RGB image with dimensions of $256^{2}$ yields a sequence length of 256. Our hierarchical Transformer incorporates three key elements to improve computational efficiency and reduce the total parameter count while maintaining the SOTA level performance. First, we incorporate overlapped patch merging to overcome neighborhood information loss during the voxel generation process. This technique, in contrast to the patching mechanism seen in ViT[7], allows the model to better understand the transition points between the voxels and has been shown to improve overall segmentation precision[27]. Next, to address the sequence length bottleneck without compromising performance, we integrate an efficient self-attention mechanism [26]. This approach enables the model to capture longrange dependencies more effectively, promoting improved s cal ability and performance. Traditional self-attention takes a sequence of vectors of shape [Batch, Sequence, Features] as input and generates 3 unique projections, Query, Key and Value vectors. Once generated, the attention scores are computed as $\begin{array}{r}{(Q,K,V)=\mathrm{Softmax}\left(\frac{Q K^{T}}{\sqrt{d_{\mathrm{head}}}}\right)V}\end{array}$ . Due to the operation $Q K^{T}$ , the computational complexity of the original segmentation process is ${\mathcal{O}}(n^{2})$ . Although this complexity can be overlooked with 2D images, with long 3D sequences, it proves to be a challenge for efficient architecture design. Efficient attention introduced in [26, 27].
编码器:在Transformer框架中使用3D医学图像会导致序列长度过长,从而增加模型的计算复杂度。例如,一个标准的128³维度的3D MRI体数据会产生32,768的序列长度,而典型的$256^{2}$维度的2D RGB图像仅产生256的序列长度。我们的分层Transformer通过三个关键要素在保持SOTA性能的同时提升计算效率并减少总参数量。首先,我们采用重叠块合并技术来克服体素生成过程中的邻域信息丢失问题。与ViT[7]中的分块机制不同,该技术使模型能更好地理解体素间的过渡点,并已被证明能提升整体分割精度[27]。其次,为在不影响性能的前提下解决序列长度瓶颈,我们整合了高效自注意力机制[26]。该方法使模型能更有效地捕捉长程依赖关系,从而提升可扩展性和性能。传统自注意力以形状为[Batch, Sequence, Features]的向量序列作为输入,生成查询(Query)、键(Key)和值(Value)三个独特投影。生成后,注意力分数通过$\begin{array}{r}{(Q,K,V)=\mathrm{Softmax}\left(\frac{Q K^{T}}{\sqrt{d_{\mathrm{head}}}}\right)V}\end{array}$计算。由于$Q K^{T}$运算的存在,原始分割过程的计算复杂度为${\mathcal{O}}(n^{2})$。虽然这种复杂度在2D图像中可以忽略,但对于长3D序列而言,它成为高效架构设计的挑战。[26,27]中引入了高效注意力机制。
$$
\begin{array}{r c l}{{\hat{K}}}&{{=}}&{{\mathrm{Reshape}(\displaystyle\frac{N}{R},C\cdot R)(K),}}\ {{}}&{{}}&{{}}\ {{K}}&{{=}}&{{\operatorname{Linear}(C\cdot R,C)(\hat{K}),}}\end{array}
$$
$$
\begin{array}{r c l}{{\hat{K}}}&{{=}}&{{\mathrm{Reshape}(\displaystyle\frac{N}{R},C\cdot R)(K),}}\ {{}}&{{}}&{{}}\ {{K}}&{{=}}&{{\operatorname{Linear}(C\cdot R,C)(\hat{K}),}}\end{array}
$$
significantly reduces the computational complexity generated by 3D volumetric tensors from ${\mathcal{O}}(n^{2})$ to $\mathcal{O}(n^{2}/r)$ . We set the reduction parameter $r$ to $4\times$ , $2\times$ , $1\times$ , $1\times$ in the four stages of the encoder.
将3D体积张量产生的计算复杂度从 ${\mathcal{O}}(n^{2})$ 显著降低至 $\mathcal{O}(n^{2}/r)$ 。我们在编码器的四个阶段将缩减参数 $r$ 分别设置为 $4\times$ 、 $2\times$ 、 $1\times$ 、 $1\times$ 。
Finally, our approach addresses the challenge of resizing volumetric imaging and its relation to fixed positional encoding in ViTs by adopting the mix-ffn module [27]. This module enables automatic learning of positional cues, elim- inating the need for fixed encoding, ensuring superior scalability and performance.
最后,我们的方法通过采用 mix-ffn 模块 [27] 解决了体积成像调整大小及其与 ViT 中固定位置编码关系的挑战。该模块能自动学习位置线索,无需固定编码,从而确保卓越的可扩展性和性能。
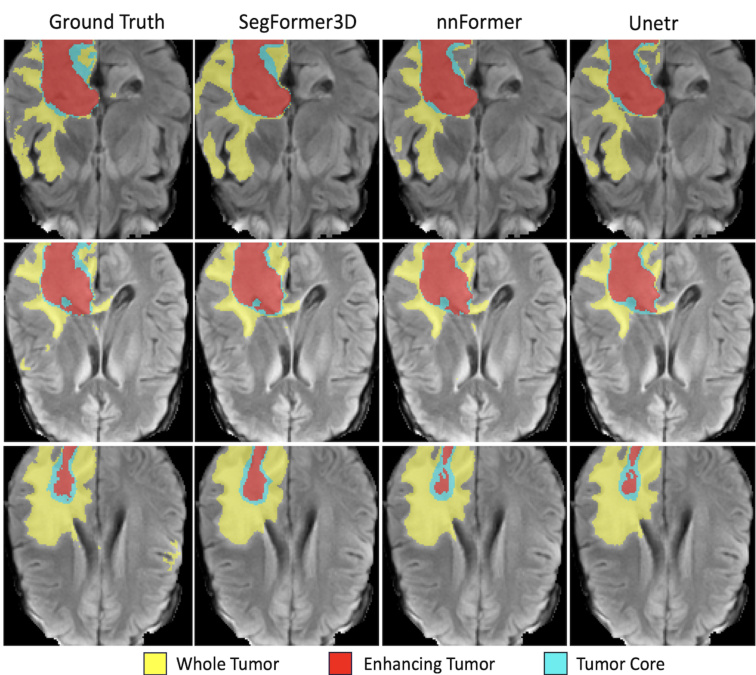
Fig. 2: Qualitative results on BRaTs. Each row is a separate frame in the MRI sequence while each column is 3D volumetric image segmentation solution. We qualitatively demonstrate highly accurate segmentation performance to SOTA methods while maintaining a lightweight and efficient architecture.
图 2: BRaTs定性结果。每行代表MRI序列中的单独帧,每列为3D体积图像分割方案。我们定性展示了与SOTA方法相比的高精度分割性能,同时保持了轻量高效的架构。
Table 2: BRaTs comparison table ranked based on average performance across all classes. Seg former 3 D is highly competitive out performing well established solutions across all categories.
| 方法 | 参数量 | 平均提升%↑ | 全肿瘤↑ | 增强肿瘤↑ | 肿瘤核心↑ |
|---|---|---|---|---|---|
| nnFormer[32] Ours | 150.5 | 86.4 | 91.3 | 81.8 | 86.0 |
| UNETR[11] | 92.49 | 71.1 | 78.9 | 74.2 | 82.2 |
| TransBTS[25] CoTr[28] | 41.9 | 69.6 | 77.9 | 57.4 | 73.5 |
| CoTrw/oCNNEncoder[28] | - | 64.4 | 71.2 | 55.7 | 74.8 |
| TransUNet[5] | 96.07 | 64.4 | 70.6 | 54.2 | 68.4 |
| SETRMLA[31] | 310.5 | 63.9 | 69.8 | 55.4 | 66.5 |
| SETRPUP[31] | 318.31 | 63.8 | 69.6 | 54.9 | 67.0 |
| SETRNUP[31] | 305.67 | 63.7 | 69.7 | 54.4 | 66.9 |
表 2: 基于各类别平均性能排序的BRaTs对比表。SegFormer 3D在所有类别中均表现出色,超越了现有成熟解决方案。
Decoder: The decoding stage plays a pivotal role in medical image segmentation based on the encoder-decoder design widely adopted in the UNET based architectures[10, 11]. This framework is used in both CNN-based and Transformer-based encoders. In the context of 3D medical images, where successive 3D convolutions are often necessary for effective decoding, we instead demonstrate that the integration of linear layers is a highly effective decoding strategy for medical image segmentation. Our approach simplifies the decoding process, ensuring efficient and consistent decoding of volumetric features across diverse datasets without over-parameter iz ation. The simple
解码器:在基于UNET架构[10,11]广泛采用的编码器-解码器设计中,解码阶段对医学图像分割起着关键作用。该框架既适用于基于CNN的编码器,也适用于基于Transformer的编码器。在处理3D医学图像时,虽然通常需要连续的3D卷积进行有效解码,但我们证明线性层的整合是一种高效的医学图像分割解码策略。我们的方法简化了解码流程,确保在不同数据集上对体积特征进行高效且一致的解码,同时避免过度参数化。
decoder process is:
解码器流程为:
$$
\begin{array}{r l}{\overset{\underset{\r}{\mathop{v s}}}{F_{i}}}&{=\mathrm{Linear}(C_{i},C)(F_{i}),\quad\forall i}\ {\hat{F}{i}}&{=\mathrm{Upsa