Learning Semantics-enriched Representation via Self-discovery, Self-classification, and Self-restoration
通过自我发现、自我分类和自我恢复学习语义增强表示
Abstract. Medical images are naturally associated with rich semantics about the human anatomy, reflected in an abundance of recurring anatomical patterns, offering unique potential to foster deep semantic representation learning and yield semantically more powerful models for different medical applications. But how exactly such strong yet free semantics embedded in medical images can be harnessed for self-supervised learning remains largely unexplored. To this end, we train deep models to learn semantically enriched visual representation by self-discovery, selfclassification, and self-restoration of the anatomy underneath medical images, resulting in a semantics-enriched, general-purpose, pre-trained 3D model, named Semantic Genesis. We examine our Semantic Genesis with all the publicly-available pre-trained models, by either self-supervision or fully supervision, on the six distinct target tasks, covering both classification and segmentation in various medical modalities (i.e., CT, MRI, and X-ray). Our extensive experiments demonstrate that Semantic Genesis significantly exceeds all of its 3D counterparts as well as the de facto ImageNet-based transfer learning in 2D. This performance is attributed to our novel self-supervised learning framework, encouraging deep models to learn compelling semantic representation from abundant anatomical patterns resulting from consistent anatomies embedded in medical images. Code and pre-trained Semantic Genesis are available at https://github.com/JLiangLab/Semantic Genesis.
摘要。医学图像天然蕴含丰富的人体解剖语义信息,通过大量重复出现的解剖模式得以体现,这为促进深度语义表征学习及构建更强大的医学应用模型提供了独特潜力。然而,如何有效利用医学图像中这种强烈且自由的语义进行自监督学习仍亟待探索。为此,我们训练深度模型通过自发现、自分类和自恢复医学图像底层解剖结构来学习语义增强的视觉表征,最终构建出名为Semantic Genesis的通用预训练3D模型。我们在六项不同目标任务(涵盖CT、MRI和X射线等多种医学模态的分类与分割)上,对所有公开可用的自监督或全监督预训练模型进行了全面评测。大量实验表明,Semantic Genesis显著优于所有3D对比模型及基于ImageNet的2D迁移学习方法。这一优势源于我们新颖的自监督学习框架,该框架促使深度模型从医学图像中固有解剖结构所产生的大量解剖模式中学习具有说服力的语义表征。代码及预训练模型详见https://github.com/JLiangLab/Semantic_Genesis。
Keywords: Self-supervised learning · Transfer learning · 3D model pretraining.
关键词:自监督学习 · 迁移学习 · 3D模型预训练
1 Introduction
1 引言
Self-supervised learning methods aim to learn general image representation from unlabeled data; naturally, a crucial question in self-supervised learning is how to “extract” proper supervision signals from the unlabeled data directly. In large part, self-supervised learning approaches involve predicting some hidden properties of the data, such as color iz ation [16,17], jigsaw [15,18], and rotation [11,13].
自监督学习方法旨在从未标注数据中学习通用的图像表征;自然,自监督学习中的一个关键问题是如何直接从无标注数据中“提取”合适的监督信号。在很大程度上,自监督学习方法涉及预测数据的某些隐藏属性,例如着色 [16,17]、拼图 [15,18] 和旋转 [11,13]。
However, most of the prominent methods were derived in the context of natural images, without considering the unique properties of medical images.
然而,大多数知名方法都是在自然图像的背景下提出的,并未考虑医学图像的特殊属性。
In medical imaging, it is required to follow protocols for defined clinical purposes, therefore generating images of similar anatomies across patients and yielding recurrent anatomical patterns across images (see Fig. 1a). These recurring patterns are associated with rich semantic knowledge about the human body, thereby offering great potential to foster deep semantic representation learning and produce more powerful models for various medical applications. However, it remains an unanswered question: How to exploit the deep semantics associated with recurrent anatomical patterns embedded in medical images to enrich representation learning?
在医学影像领域,需要遵循针对特定临床目的制定的协议,因此能生成不同患者间的相似解剖结构图像,并在图像中呈现重复出现的解剖模式 (见图 1a)。这些重复模式与人体丰富的语义知识相关联,从而为促进深度语义表征学习提供了巨大潜力,并能构建更强大的模型以应用于各类医疗场景。然而,如何利用医学影像中重复解剖模式所蕴含的深层语义来丰富表征学习,仍是一个悬而未决的问题。
To answer this question, we present a novel self-supervised learning framework, which enables the capture of semantics-enriched representation from unlabeled medical image data, resulting in a set of powerful pre-trained models. We call our pre-trained models Semantic Genesis, because they represent a significant advancement from Models Genesis [25] by introducing two novel components: self-discovery and self-classification of the anatomy underneath medical images (detailed in Sec. 2). Specifically, our unique self-classification branch, with a small computational overhead, compels the model to learn semantics from consistent and recurring anatomical patterns discovered during the selfdiscovery phase, while Models Genesis learns representation from random subvolumes with no semantics as no semantics can be discovered from random sub-volumes. By explicitly employing the strong yet free semantic supervision signals, Semantic Genesis distinguishes itself from all other existing works, including color iz ation of colon os copy images [20], context restoration [9], Rubik’s cube recovery [26], and predicting anatomical positions within MR images [4].
为回答这一问题,我们提出了一种新颖的自监督学习框架,能够从未标注的医学图像数据中捕获富含语义的表征,从而生成一组强大的预训练模型。我们将这些预训练模型命名为Semantic Genesis,因为它们通过引入两个创新组件(详见第2节)——医学图像底层解剖结构的自发现(self-discovery)与自分类(self-classification),实现了对Models Genesis [25] 的重大改进。具体而言,我们独特的自分类分支以极小的计算开销,迫使模型从自发现阶段识别出的、具有一致性和重复性的解剖模式中学习语义,而Models Genesis仅从无语义的随机子体积中学习表征(因为随机子体积无法发现任何语义)。通过显式利用这种强大且免费的语义监督信号,Semantic Genesis与所有现有工作形成鲜明区分,包括结肠镜图像着色 [20]、上下文恢复 [9]、魔方复原 [26] 以及MR图像内解剖位置预测 [4]。
As evident in Sec. 4, our extensive experiments demonstrate that (1) learning semantics through our two innovations significantly enriches existing selfsupervised learning approaches [9,19,25], boosting target tasks performance dramatically (see Fig. 2); (2) Semantic Genesis provides more generic and transferable feature representations in comparison to not only its self-supervised learning counterparts, but also (fully) supervised pre-trained 3D models (see Table 2); and Semantic Genesis significantly surpasses any 2D approaches (see Fig. 3).
如第4节所示,我们的大量实验证明:(1) 通过我们的两项创新学习语义显著丰富了现有的自监督学习方法 [9,19,25],大幅提升了目标任务性能 (见图2);(2) 与自监督学习方案及(完全)监督预训练的3D模型相比,Semantic Genesis能提供更通用、可迁移性更强的特征表示 (见表2);同时Semantic Genesis显著超越了所有2D方案 (见图3)。
This performance is ascribed to the semantics derived from the consistent and recurrent anatomical patterns, that not only can be automatically discovered from medical images but can also serve as strong yet free supervision signals for deep models to learn more semantically enriched representation automatically via self-supervision.
这一性能归功于从一致且反复出现的解剖模式中提取的语义信息,这些模式不仅能从医学图像中自动发现,还能作为强大而自由的监督信号,使深度学习模型通过自监督自动学习更具语义丰富性的表征。
2 Semantic Genesis
2 语义起源
Fig. 1 presents our self-supervised learning framework, which enables training Semantic Genesis from scratch on unlabeled medical images. Semantic Genesis is conceptually simple: an encoder-decoder structure with skip connections in between and a classification head at the end of the encoder. The objective for the model is to learn different sets of semantics-enriched representation from multiple perspectives. In doing so, our proposed framework consists of three important components: 1) self-discovery of anatomical patterns from similar patients; 2) self-classification of the patterns; and 3) self-restoration of the transformed patterns. Specifically, once the self-discovered anatomical pattern set is built, we jointly train the classification and restoration branches together in the model.
图 1 展示了我们的自监督学习框架,该框架能够在未标记的医学图像上从头开始训练 Semantic Genesis。Semantic Genesis 在概念上很简单:一个编码器-解码器结构,中间有跳跃连接,编码器末端有一个分类头。该模型的目标是从多个角度学习不同的语义丰富表示集。为此,我们提出的框架包含三个重要组成部分:1) 从相似患者中自我发现解剖模式;2) 对模式进行自分类;3) 对转换后的模式进行自我恢复。具体来说,一旦建立了自我发现的解剖模式集,我们就在模型中联合训练分类和恢复分支。
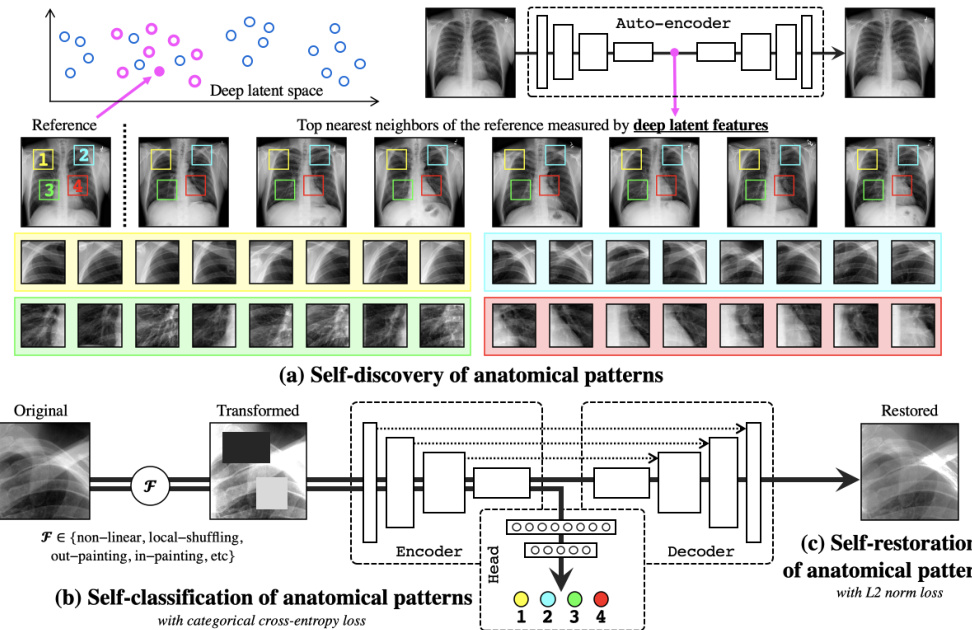
Fig. 1. Our self-supervised learning framework consists of (a) self-discovery, (b) selfclassification, and (c) self-restoration of anatomical patterns, resulting in semanticsenriched pre-trained models—Semantic Genesis—an encoder-decoder structure with skip connections in between and a classification head at the end of the encoder. Given a random reference patient, we find similar patients based on deep latent features, crop anatomical patterns from random yet fixed coordinates, and assign pseudo labels to the crops according to their coordinates. For simplicity and clarity, we illustrate our idea with four coordinates in X-ray images as an example. The input to the model is a transformed anatomical pattern crop, and the model is trained to classify the pseudo label and to recover the original crop. Thereby, the model aims to acquire semanticsenriched representation, producing more powerful application-specific target models.
图 1: 我们的自监督学习框架包含 (a) 自发现、(b) 自分类和 (c) 解剖模式自恢复三个模块,最终形成语义增强的预训练模型——Semantic Genesis。该模型采用编码器-解码器结构,中间带有跳跃连接,编码器末端配有分类头。给定随机参考患者后,我们基于深度潜在特征寻找相似患者,从随机但固定的坐标位置裁剪解剖模式,并根据坐标位置为裁剪区域分配伪标签。为简明起见,我们以X光图像中的四个坐标为例进行说明。模型输入是经过变换的解剖模式裁剪区域,训练目标是分类伪标签并重建原始裁剪区域。通过这种方式,模型旨在获得语义增强的表征,从而生成更强大的应用专用目标模型。
- Self-discovery of anatomical patterns: We begin by building a set of anatomical patterns from medical images, as illustrated in Fig. 1a. To extract deep features of each (whole) patient scan, we first train an auto-encoder network with training data, which learns an identical mapping from scan to itself. Once trained, the latent representation vector from the auto-encoder can be used as an indicator of each patient. We randomly anchor one patient as a reference and search for its nearest neighbors through the entire dataset by computing the $L2$ distance of the latent representation vectors, resulting in a set of semantically similar patients. As shown in Fig. 1a, due to the consistent and recurring anatomies across these patients, that is, each coordinate contains a unique anatomical pattern, it is feasible to extract similar anatomical patterns according to the coordinates. Hence, we crop patches/cubes (for 2D/3D images) from $C$ number of random but fixed coordinates across this small set of discovered patients, which share similar semantics. Here we compute similarity in patient-level rather than pattern-level to ensure the balance between the diversity and consistency of anatomical patterns. Finally, we assign pseudo labels to these patches/cubes based on their coordinates, resulting in a new dataset, wherein each patch/cube is associated with one of the $C$ classes. Since the coordinates are randomly selected in the reference patient, some of the anatomical patterns may not be very meaningful for radiologists, yet these patterns are still associated with rich local semantics of the human body. For example, in Fig. 1a, four pseudo labels are defined randomly in the reference patient (top-left most), but as seen, they carry local information of (1) anterior ribs 2–4, (2) anterior ribs 1–3, (3) right pulmonary artery, and (4) LV. Most importantly, by repeating the above self-discovery process, enormous anatomical patterns associated with their pseudo labels can be automatically generated for representation learning in the following stages (refer to Appendix Sec. A).
- 解剖模式自发现:我们首先从医学图像中构建一组解剖模式,如图1a所示。为提取每例(完整)患者扫描的深层特征,我们先用训练数据训练一个自编码器网络,该网络学习从扫描到自身的恒等映射。训练完成后,自编码器的潜在表示向量可作为每位患者的表征指标。随机选取一位患者作为参考锚点,通过计算潜在表示向量的$L2$距离在整个数据集中搜索其最近邻,从而获得一组语义相似的患者。如图1a所示,由于这些患者具有一致且重复出现的解剖结构(即每个坐标点对应独特的解剖模式),按坐标提取相似解剖模式具有可行性。因此,我们从这组语义相似的已发现患者中,在$C$个随机但固定的坐标位置上裁剪图像块/立方体(分别对应2D/3D图像)。此处采用患者级而非模式级的相似度计算,以确保解剖模式多样性与一致性的平衡。最后根据坐标位置为这些图像块/立方体分配伪标签,形成新数据集,其中每个图像块/立方体归属于$C$个类别之一。由于坐标是在参考患者中随机选取的,部分解剖模式对放射科医生可能意义不大,但这些模式仍关联着丰富的人体局部语义。例如图1a中,参考患者(左上角)随机定义了四个伪标签,实际分别对应:(1) 第2-4前肋,(2) 第1-3前肋,(3) 右肺动脉,(4) 左心室的局部信息。最重要的是,通过重复上述自发现过程,可自动生成海量带伪标签的解剖模式用于后续阶段的表征学习(详见附录A节)。
- Self-classification of anatomical patterns: After self-discovery of a set of anatomical patterns, we formulate the representation learning as a $C$ -way multi-class classification task. The goal is to encourage models to learn from the recurrent anatomical patterns across patient images, fostering a deep semantically enriched representation. As illustrated in Fig. 1b, the classification branch encodes the input anatomical pattern into a latent space, followed by a sequence of fully-connected $(f c)$ layers, and predicts the pseudo label associated with the pattern. To classify the anatomical patterns, we adopt categorical cross-entropy loss function: $\begin{array}{r}{\mathcal{L}{c l s}=-\frac{1}{N}\sum_{b=1}^{N}\sum_{c=1}^{C}\mathcal{V}{b c}\log\mathcal{P}_{b c}}\end{array}$ , where $N$ denotes the batch size; $C$ denotes the number of classes; $\mathcal{V}$ and $\mathcal{P}$ represent the ground truth (one-hot pseudo label vector) and the prediction, respectively.
- 解剖模式自分类: 在自主发现一组解剖模式后,我们将表征学习建模为一个$C$类多分类任务。该目标旨在促使模型从患者图像中反复出现的解剖模式中学习,从而形成深度语义增强的表征。如图1b所示,分类分支将输入解剖模式编码至潜在空间,随后通过全连接层$(fc)$序列,预测与该模式关联的伪标签。我们采用分类交叉熵损失函数进行解剖模式分类: $\begin{array}{r}{\mathcal{L}{c l s}=-\frac{1}{N}\sum_{b=1}^{N}\sum_{c=1}^{C}\mathcal{V}{b c}\log\mathcal{P}_{b c}}\end{array}$,其中$N$表示批次大小,$C$表示类别数量,$\mathcal{V}$和$\mathcal{P}$分别代表真实值(独热伪标签向量)和预测值。
- Self-restoration of anatomical patterns: The objective of self-restoration is for the model to learn different sets of visual representation by recovering original anatomical patterns from the transformed ones. We adopt the transformations proposed in Models Genesis [25], i.e., non-linear, local-shuffling, outpainting, and in-painting (refer to Appendix Sec. B). As shown in Fig. 1c, the restoration branch encodes the input transformed anatomical pattern into a latent space and decodes back to the original resolution, with an aim to recover the original anatomical pattern from the transformed one. To let Semantic Genesis restore the transformed anatomical patterns, we compute $L2$ distance between original pattern and reconstructed pattern as loss function: $\begin{array}{r}{\mathcal{L}{r e c}=\frac{1}{N}\sum_{i=1}^{N}|\mathcal{X}{i}-\mathcal{X}{i}^{\prime}|_{2}}\end{array}$ , where $N$ , $\mathcal{X}$ and $\mathcal{X}^{\prime}$ denote the batch size, ground truth (original anatomical pattern) and reconstructed prediction, respectively.
- 解剖模式自恢复:自恢复的目标是让模型通过从变换后的解剖模式中恢复原始解剖模式,学习不同的视觉表示集。我们采用Models Genesis [25]提出的变换方法,即非线性变换、局部打乱、外绘和内绘(详见附录B节)。如图1c所示,恢复分支将输入的变换解剖模式编码到潜在空间,并解码回原始分辨率,旨在从变换后的模式中恢复原始解剖模式。为使Semantic Genesis能恢复变换后的解剖模式,我们计算原始模式与重建模式之间的$L2$距离作为损失函数:$\begin{array}{r}{\mathcal{L}{r e c}=\frac{1}{N}\sum_{i=1}^{N}|\mathcal{X}{i}-\mathcal{X}{i}^{\prime}|_{2}}\end{array}$,其中$N$、$\mathcal{X}$和$\mathcal{X}^{\prime}$分别表示批次大小、真实值(原始解剖模式)和重建预测结果。
Formally, during training, we define a multi-task loss function on each transformed anatomical pattern as $\mathcal{L}=\lambda_{c l s}\mathcal{L}{c l s}+\lambda_{r e c}\mathcal{L}{r e c}$ , where $\lambda_{c l s}$ and $\lambda_{r e c}$ regulate the weights of classification and reconstruction losses, respectively. Our definition of $\mathcal{L}{c l s}$ allows the model to learn more semantically enriched representation. The definition of $\mathscr{L}{r e c}$ encourages the model to learn from multiple perspectives by restoring original images from varying image deformations. Once trained, the encoder alone can be fine-tuned for target classification tasks; while the encoder and decoder together can be fine-tuned for target segmentation tasks to fully utilize the advantages of the pre-trained models on the target tasks.
在训练过程中,我们为每个变换后的解剖模式定义了一个多任务损失函数 $\mathcal{L}=\lambda_{c l s}\mathcal{L}{c l s}+\lambda_{r e c}\mathcal{L}{r e c}$ ,其中 $\lambda_{c l s}$ 和 $\lambda_{r e c}$ 分别调节分类损失和重建损失的权重。$\mathcal{L}{c l s}$ 的定义使模型能够学习语义更丰富的表示,而 $\mathscr{L}_{r e c}$ 的定义则通过从不同图像变形中恢复原始图像,促使模型从多角度学习。训练完成后,可单独对编码器进行微调以适配目标分类任务;同时微调解码器与编码器则可应用于目标分割任务,从而充分利用预训练模型在目标任务中的优势。
Table 1. We evaluate the learned representation by fine-tuning it for six publiclyavailable medical imaging applications including 3D and 2D image classification and segmentation tasks, across diseases, organs, datasets, and modalities.
a The first letter denotes the object of interest (“N” for lung nodule, “L” for liver, etc); the second letter denotes the modality (“C” for CT, “X” for X-ray, “M” for MRI); the last letter denotes the task (“C” for classification, “S” for segmentation).
表 1: 我们通过微调学习到的表征,评估了其在六种公开可用的医学影像应用中的表现,涵盖3D和2D图像分类与分割任务,涉及不同疾病、器官、数据集和模态。
| 代码对象 | 目标部位 | 模态 | 数据集 | 应用 |
|---|---|---|---|---|
| NCC | 肺结节 | CT | LUNA-2016 [21] | 结节假阳性减少 |
| NCS | 肺结节 | CT | LIDC-IDRI [3] | 肺结节分割 |
| LCS | 肝脏 | CT | LiTS-2017 [6] | 肝脏分割 |
| BMS | 脑肿瘤 | MRI | BraTS2018 [5] | 脑肿瘤分割 |
| DXC | 胸部疾病 | X-ray | ChestX-Ray14 [23] | 十四种胸部疾病分类 |
| PXS | 气胸 | X-ray | SIIM-ACR-2019 [1] | 气胸分割 |
a 首字母表示目标部位 ("N"代表肺结节,"L"代表肝脏等);第二个字母表示模态 ("C"代表CT,"X"代表X射线,"M"代表MRI);最后一个字母表示任务 ("C"代表分类,"S"代表分割)。
3 Experiments
3 实验
Pre-training Semantic Genesis: Our Semantic Genesis 3D and 2D are selfsupervised pre-trained from 623 CT scans in LUNA-2016 [21] (same as the publicly released Models Genesis) and 75,708 X-ray images from ChestX-ray14 [22] datasets, respectively. Although Semantic Genesis is trained from only unlabeled images, we do not use all the images in those datasets to avoid test-image leaks between proxy and target tasks. In the self-discovery process, we select top $K$ most similar cases with the reference patient, according to the deep features computed from the pre-trained auto-encoder. To strike a balance between diversity and consistency of the anatomical patterns, we empirically set $K$ to 200/1000 for 3D/2D pre-training based on the dataset size. We set $C$ to 44/100 for 3D/2D images so that the anatomical patterns can largely cover the entire image while avoiding too much overlap with each other. For each random coordinate, we extract multi-resolution cubes/patches, then resize them all to 64 $\times$ 64 $\times$ 32 and 224 $\times$ 224 for 3D and 2D, respectively; finally, we assign $C$ pseudo labels to the cubes/patches based on their coordinates. For more details in implementation and meta-parameters, please refer to our publicly released code.
预训练语义起源:我们的Semantic Genesis 3D和2D模型分别通过自监督方式预训练,数据来源为LUNA-2016 [21] 数据集中的623例CT扫描(与公开发布的Models Genesis相同)和ChestX-ray14 [22] 数据集中的75,708张X光片。尽管语义起源仅使用未标注图像进行训练,但为避免代理任务与目标任务间的测试图像泄露,我们并未使用这些数据集中的所有图像。在自发现过程中,我们根据预训练自编码器提取的深度特征,筛选出与参考患者最相似的$K$个病例。为平衡解剖模式的多样性与一致性,基于数据集规模,我们经验性地将3D/2D预训练的$K$值设为200/1000。将$C$设置为44/100(3D/2D图像),确保解剖模式能覆盖大部分图像区域,同时避免过多重叠。针对每个随机坐标,我们提取多分辨率立方体/图像块,并将其统一调整为64$\times$64$\times$32(3D)和224$\times$224(2D),最后根据坐标位置为立方体/图像块分配$C$个伪标签。具体实现细节与元参数请参阅我们公开的代码。
Baselines and implementation: Table 1 summarizes the target tasks and datasets. Since most self-supervised learning methods are initially proposed in 2D, we have extended two most representative ones [9,19] into their 3D version for a fair comparison. Also, we compare Semantic Genesis with Rubik’s cube [26], the most recent multi-task self-supervised learning method for 3D medical imaging. In addition, we have examined publicly available pre-trained models for 3D transfer learning in medical imaging, including NiftyNet [12], MedicalNet [10], Models Genesis [25], and Inflated 3D (I3D) [8] that has been successfully transferred to 3D lung nodule detection [2], as well as ImageNet models, the most influential weights initialization in 2D target tasks. 3D U-Net $^{3}$ /U-Net $^4$ architectures used in 3D/2D applications, have been modified by appending fullyconnected layers to end of the encoders. In proxy tasks, we set $\lambda_{r e c}=1$ and $\lambda_{c l s}~=~0.01$ . Adam with a learning rate of 0.001 is used for optimization. We first train classification branch for 20 epochs, then jointly train the entire model for both classification and restoration tasks. For CT target tasks, we investigate the capability of both 3D volume-based solutions and 2D slice-based solutions, where the 2D representation is obtained by extracting axial slices from volumetric datasets. For all applications, we run each method 10 times on the target task and report the average, standard deviation, and further present statistical analyses based on independent two-sample $t$ -test.
基准方法与实现:表1总结了目标任务和数据集。由于大多数自监督学习方法最初是针对2D提出的,我们扩展了两种最具代表性的方法[9,19]到3D版本以确保公平比较。同时,我们将Semantic Genesis与最新的3D医学影像多任务自监督学习方法Rubik's cube[26]进行对比。此外,我们还评估了医学影像3D迁移学习的公开预训练模型,包括NiftyNet[12]、MedicalNet[10]、Models Genesis[25]以及已成功迁移至3D肺结节检测的Inflated 3D (I3D)[8],还有在2D目标任务中最具影响力的ImageNet模型权重初始化方案。针对3D/2D应用场景的3D U-Net$^{3}$/U-Net$^4$架构,我们通过在编码器末端添加全连接层进行了修改。在代理任务中,设置$\lambda_{r e c}=1$和$\lambda_{c l s}~=~0.01$,采用学习率为0.001的Adam优化器。先单独训练分类分支20个周期,再联合训练整个模型完成分类和重建任务。对于CT目标任务,我们同时研究了基于3D体素和2D切片的解决方案,其中2D表示通过从体数据集中提取轴向切片获得。所有实验均在目标任务上重复运行10次,报告平均值、标准差,并基于独立双样本$t$检验进行统计分析。
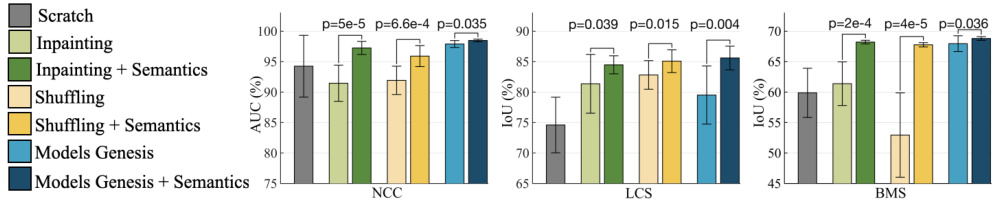
Fig. 2. With and without semantics-enriched representation in the self-supervised learning approaches contrast a substantial ( $\textit{p}<0.05)$ performance difference on target classification and segmentation tasks. By introducing self-discovery and selfclassification, we enhance semantics in three most recent self-supervised learning advances (i.e., image in-painting [19], patch-shuffling [9], and Models Genesis [25]).
图 2: 在自监督学习方法中,使用和未使用语义增强表示对目标分类和分割任务性能存在显著差异 (p<0.05)。通过引入自发现和自分类机制,我们在三种前沿自监督学习方法 (即图像修复 [19]、块重排 [9] 和 Models Genesis [25]) 中增强了语义表征能力。
