G-CASCADE: Efficient Cascaded Graph Convolutional Decoding for 2D Medical Image Segmentation
G-CASCADE: 用于二维医学图像分割的高效级联图卷积解码
Abstract
摘要
In recent years, medical image segmentation has become an important application in the field of computer-aided diagnosis. In this paper, we are the first to propose a new graph convolution-based decoder namely, Cascaded Graph Convolutional Attention Decoder (G-CASCADE), for $2D$ medical image segmentation. G-CASCADE progressively refines multi-stage feature maps generated by hierarchical transformer encoders with an efficient graph convolution block. The encoder utilizes the self-attention mechanism to capture long-range dependencies, while the decoder refines the feature maps preserving long-range information due to the global receptive fields of the graph convolution block. Rigorous evaluations of our decoder with multiple transformer encoders on five medical image segmentation tasks (i.e., Abdomen organs, Cardiac organs, Polyp lesions, Skin lesions, and Retinal vessels) show that our model outperforms other state-of-the-art (SOTA) methods. We also demonstrate that our decoder achieves better DICE scores than the SOTA CASCADE decoder with $80.8%$ fewer parameters and $82.3%$ fewer FLOPs. Our decoder can easily be used with other hierarchical encoders for generalpurpose semantic and medical image segmentation tasks.
近年来,医学图像分割已成为计算机辅助诊断领域的重要应用。本文首次提出了一种基于图卷积的新型解码器——级联图卷积注意力解码器 (G-CASCADE) ,用于 $2D$ 医学图像分割。G-CASCADE通过高效的图卷积模块逐步细化由分层Transformer编码器生成的多阶段特征图。编码器利用自注意力机制捕获长程依赖关系,而解码器则借助图卷积模块的全局感受野保留长程信息来优化特征图。我们在五项医学图像分割任务 (即腹部器官、心脏器官、息肉病变、皮肤病变和视网膜血管) 中,使用多种Transformer编码器对解码器进行严格评估,结果表明该模型性能优于其他最先进 (SOTA) 方法。我们还证明,与SOTA CASCADE解码器相比,该解码器以减少 $80.8%$ 参数量和 $82.3%$ FLOPs的代价获得了更高的DICE分数。该解码器可轻松适配其他分层编码器,适用于通用语义分割及医学图像分割任务。
1. Introduction
1. 引言
and improve pixel-level classification of medical images by capturing salient features. Although these attentionbased methods have shown improved performance, they still struggle to capture long-range dependencies [28].
并通过捕捉显著特征来提升医学图像的像素级分类效果。尽管这些基于注意力机制的方法已展现出性能提升,但它们仍难以捕捉长距离依赖关系 [28]。
Recently, vision transformers [10] has shown great promise in capturing long-range dependencies among pixels and demonstrated improved performance, particularly for medical image segmentation [4, 2, 9, 38, 28, 29, 48, 36]. The self-attention (SA) mechanism used in transformers learns correlations between input patches; this enables capturing the long-range dependencies among pixels. Recently, hierarchical vision transformers such as the Swin transformer [23], the pyramid vision transformer (PVT) [39], MaxViT [34], MERIT [29], have been introduced to enhance performance. These hierarchical vision transformers are effective in medical image segmentation tasks [4, 2, 9, 38, 28, 29]. As self-attention modules employed in transformers have limited capacity to learn (local) spatial relationships among pixels [7, 17], some methods [44, 42, 40, 9, 38, 28, 29] incorporate local convolutional attention modules in the decoder. However, due to the locality of convolution operations, these methods have difficulties at capturing long-range correlations among pixels.
近年来,视觉Transformer (vision transformers) [10] 在捕捉像素间长程依赖关系方面展现出巨大潜力,并在医学图像分割任务中表现出优越性能 [4, 2, 9, 38, 28, 29, 48, 36]。Transformer中的自注意力 (self-attention, SA) 机制通过学习输入图像块之间的相关性,能够有效建模像素间的长程依赖。近期提出的分层视觉Transformer——如Swin Transformer [23]、金字塔视觉Transformer (PVT) [39]、MaxViT [34] 和 MERIT [29]——进一步提升了性能表现,这些结构在医学图像分割任务中表现优异 [4, 2, 9, 38, 28, 29]。由于Transformer的自注意力模块在学习像素间 (局部) 空间关系时存在局限性 [7, 17],部分方法 [44, 42, 40, 9, 38, 28, 29] 在解码器中引入了局部卷积注意力模块。然而,受限于卷积操作的局部性,这些方法仍难以有效捕捉像素间的长程关联。
Automatic medical image segmentation plays a crucial role in the diagnosis, treatment planning, and post-treatment evaluation of various diseases; this involves classifying pixels and generating segmentation maps to identify lesions, tumours, or organs. Convolutional neural networks (CNNs) have been extensively utilized for medical image segmentation tasks [30, 27, 49, 15, 11, 26]. Among them, the Ushaped networks such as UNet [30], $\mathrm{UNet}++$ [49], UNet $^{3+}$ [15], and DC-UNet [26] exhibit reasonable performance and produce high-resolution segmentation maps. Additionally, researchers have incorporated attention modules into their architectures [27, 6, 11] to enhance feature maps
自动医学图像分割在各种疾病的诊断、治疗规划和治疗后评估中起着关键作用;这涉及对像素进行分类并生成分割图以识别病变、肿瘤或器官。卷积神经网络 (CNN) 已被广泛应用于医学图像分割任务 [30, 27, 49, 15, 11, 26]。其中,U形网络如 UNet [30]、$\mathrm{UNet}++$ [49]、UNet$^{3+}$ [15] 和 DC-UNet [26] 表现出合理的性能并生成高分辨率分割图。此外,研究人员还在其架构中加入了注意力模块 [27, 6, 11] 以增强特征图。
To overcome the aforementioned limitations, we introduce a new Graph based CAScaded Convolutional Attention DEcoder (G-CASCADE) using graph convolutions. More precisely, G-CASCADE enhances the feature maps by preserving long-range attention due to the global receptive field of the graph convolution operation, while incorporating local attention through the spatial attention mechanism. Our contributions are as follows:
为克服上述局限性,我们提出了一种基于图卷积的新型图级联卷积注意力解码器 (G-CASCADE) 。具体而言,G-CASCADE通过图卷积操作的全局感受野保留长程注意力,同时结合空间注意力机制引入局部注意力,从而增强特征图。我们的贡献如下:
• New Graph Convolutional Decoder: We introduce a new graph-based cascaded convolutional attention decoder (G-CASCADE) for 2D medical image segmentation; this takes the multi-stage features of vision transformers and learns multiscale and multi resolution spatial representations. To the best of our knowledge, we are the first to propose this graph convolutional network-based decoder for semantic segmentation.
• 新图卷积解码器:我们提出了一种基于图的新型级联卷积注意力解码器 (G-CASCADE) 用于2D医学图像分割,该解码器利用视觉Transformer (Vision Transformer) 的多阶段特征,学习多尺度和多分辨率空间表示。据我们所知,这是首个基于图卷积网络的语义分割解码器。
• Efficient Graph Convolutional Attention Block: We introduce a new graph convolutional attention module to build our decoder; this preserves the long-range attention of the vision transformer and highlights salient features by suppressing irrelevant regions. The use of graph convolution makes our decoder efficient.
• 高效图卷积注意力块 (Efficient Graph Convolutional Attention Block) : 我们提出了一种新的图卷积注意力模块来构建解码器,该模块保留了视觉Transformer的长程注意力特性,并通过抑制无关区域来突出显著特征。图卷积的使用使我们的解码器更加高效。
• Efficient Design of Up-Convolution Block: We design an efficient up-convolution block that enables computational gains without degrading performance.
• 高效上采样卷积块设计:我们设计了一种高效的上采样卷积块,在不降低性能的前提下实现计算效率提升。
• Improved Performance: We empirically show that G-CASCADE can be used with any hierarchical vision encoder (e.g., PVT [40], MERIT [4]) while significantly improving the performance of 2D medical image segmentation. When compared against multiple baselines, G-CASCADE produces better results than SOTA methods on ACDC, Synapse Multi-organ, ISIC2018 skin lesion, Polyp, and Retinal vessels segmentation benchmarks with a significantly lower computational cost.
• 性能提升:我们通过实验证明,G-CASCADE可与任何分层视觉编码器(如PVT [40]、MERIT [4])配合使用,同时显著提升2D医学图像分割性能。在ACDC、Synapse多器官、ISIC2018皮肤病变、息肉及视网膜血管分割基准测试中,G-CASCADE以显著更低的计算成本,相比多种基线方法产生了优于当前最优(SOTA)方法的结果。
The remaining of this paper is organized as follows: Section 2 summarizes the related work in vision transformers, graph convolutional networks, and medical image segmentation. Section 3 describes the proposed method Section 4 explains experimental setup and results on multiple medical image segmentation benchmarks. Section 5 covers different ablation experiments. Lastly, Section 6 concludes the paper.
本文剩余部分结构如下:第2节总结了视觉Transformer、图卷积网络和医学图像分割的相关工作。第3节描述了所提出的方法。第4节阐述了多个医学图像分割基准的实验设置和结果。第5节涵盖了不同的消融实验。最后,第6节对全文进行总结。
2. Related Work
2. 相关工作
We divide the related work into three parts, i.e., vision transformers, vision graph convolutional networks, and medical image segmentation; these are described next.
我们将相关工作分为三个部分,即视觉Transformer (Vision Transformer)、视觉图卷积网络 (Vision Graph Convolutional Network) 和医学图像分割,接下来将分别进行描述。
2.1. Vision transformers
2.1. Vision transformers
Do sov it ski y et al. [10] pioneered the development of the vision transformer (ViT), which enables the learning of long-range relationships between pixels through selfattention. Subsequent works have focused on enhancing ViT in various ways, such as integration of convolutional neural networks (CNNs) [40, 34], introducing new SA blocks [23, 34], and novel architectural designs [39, 44]. Liu et al. [23] introduce a sliding window attention mechanism within the hierarchical Swin transformer. Xie et al. [44] present SegFormer, a hierarchical transformer utilizing Mix-FFN blocks. Wang et al. [39] develop the pyramid vision transformer (PVT) with a spatial reduction attention mechanism, and subsequently extend it to PVTv2 [40] by incorporating overlapping patch embedding, a linear complexity attention layer, and a convolutional feedforward network. Most recently, Tu et al. [34] introduce MaxViT, which employs a multi-axis self-attention mechanism to construct a hierarchical CNN-transformer encoder.
Do sov it ski y 等人 [10] 开创性地开发了视觉Transformer (ViT),通过自注意力机制学习像素间的长程关系。后续研究从多个方向改进ViT,例如整合卷积神经网络 (CNNs) [40, 34]、设计新型自注意力模块 [23, 34] 以及创新架构设计 [39, 44]。Liu 等人 [23] 在分层Swin transformer中引入滑动窗口注意力机制。Xie 等人 [44] 提出SegFormer,这是一种采用Mix-FFN模块的分层Transformer。Wang 等人 [39] 开发了具有空间缩减注意力机制的金字塔视觉Transformer (PVT),随后通过整合重叠块嵌入、线性复杂度注意力层和卷积前馈网络,将其扩展为PVTv2 [40]。最近,Tu 等人 [34] 提出MaxViT,采用多轴自注意力机制构建分层CNN-Transformer编码器。
Although vision transformers exhibit remarkable performance, they have certain limitations in their (local) spatial information processing capabilities. In this paper, we aim to overcome these limitations by introducing a new graphbased cascaded attention decoder that preserves the longrange attention through graph convolution and incorporates local attention by a spatial attention mechanism.
尽管视觉Transformer表现出卓越的性能,但其在(局部)空间信息处理能力上存在一定局限。本文提出一种基于图结构的级联注意力解码器,通过图卷积保持长程注意力,并引入空间注意力机制融合局部注意力。
2.2. Vision graph convolutional networks
2.2. 视觉图卷积网络
Graph convolutional networks (GCNs) are developed primarily focusing on point clouds classification [20, 21], scene graph generation [45], and action recognition [47] in computer vision. Vision GNN (ViG) [13] introduces the first graph convolutional backbone network to directly process the image data. ViG devides the image into patches and then uses K-nearest neighbors (KNN) algorithm to connect various patches; this enables the processing of long-range dependencies similar to vision transformers. Besides, due to using $1\times1$ convolutions before and after the graph convolution operation, the graph convolution block used in ViG is significantly faster than the vision transformer and $3\times3$ convolution-based CNN blocks. Therefore, we propose to use the graph convolution block to decode feature maps for dense prediction. This will make our decoder computationally efficient, while preserving long-range information.
图卷积网络 (GCN) 主要应用于计算机视觉领域的点云分类 [20, 21]、场景图生成 [45] 和动作识别 [47]。视觉图卷积网络 (ViG) [13] 首次提出了直接处理图像数据的图卷积主干网络。ViG 将图像分割为多个小块 (patch),然后使用 K 近邻 (KNN) 算法连接这些小块,从而实现了类似于视觉 Transformer 的远距离依赖处理能力。此外,由于在图卷积操作前后使用了 $1\times1$ 卷积,ViG 采用的图卷积块在速度上显著优于视觉 Transformer 和基于 $3\times3$ 卷积的 CNN 模块。因此,我们提出使用图卷积块对特征图进行解码以实现密集预测。这种方法将使解码器在保持远距离信息的同时具备更高的计算效率。
2.3. Medical image segmentation
2.3. 医学图像分割
Medical image segmentation is the task of classifying pixels into lesions, tumours, or organs in a medical image (e.g., endoscopy, MRI, and CT) [4]. To address this task, U-shaped architectures [30, 27, 49, 15, 26] have been commonly utilized due to their sophisticated encoder-decoder structure. Ronne berger et al. [15] introduce UNet, an encoder-decoder architecture that utilizes skip connections to aggregate features from multiple stages. In $\mathrm{UNet}++$ [49], nested encoder-decoder sub-networks are connected through dense skip connections. UNet $^{3+}$ [15] further extends this concept by exploring full-scale skip connections with intra-connections among the decoder blocks. DCUNet [26] incorporates the multi-resolution convolution block and residual path within skip connections. These archi tec ture s have proven to be effective in medical image segmentation tasks.
医学图像分割的任务是将医学图像(如内窥镜、MRI和CT)中的像素分类为病变、肿瘤或器官 [4]。为解决这一任务,U型架构 [30, 27, 49, 15, 26] 因其复杂的编码器-解码器结构而被广泛采用。Ronneberger等人 [15] 提出了UNet,这是一种利用跳跃连接聚合多阶段特征的编码器-解码器架构。在$\mathrm{UNet}++$ [49] 中,嵌套的编码器-解码器子网络通过密集跳跃连接相连。UNet$^{3+}$ [15] 进一步扩展了这一概念,探索了解码器块间全尺度跳跃连接与内部连接。DCUNet [26] 在跳跃连接中引入了多分辨率卷积块和残差路径。这些架构已被证明在医学图像分割任务中非常有效。
Recently, transformers have gained popularity in the field of medical image segmentation [2, 4, 9, 28, 29, 36, 48]. In TransUNet [4], a hybrid architecture combining CNNs and transformers is proposed to capture both local and global pixel relationships. Swin-Unet [2] adopts a pure Ushaped transformer structure by utilizing Swin transformer blocks [23] in both the encoder and decoder. More recently, Rahman et al. [29] propose a multi-scale hierarchical transformer network with cascaded attention decoding (MERIT) that calculates self attention in varying window sizes to capture effective multi-scale features.
最近,Transformer在医学图像分割领域越来越受欢迎[2, 4, 9, 28, 29, 36, 48]。在TransUNet[4]中,提出了一种结合CNN和Transformer的混合架构,以捕捉局部和全局像素关系。Swin-Unet[2]通过在编码器和解码器中使用Swin Transformer块[23],采用了一种纯U形Transformer结构。最近,Rahman等人[29]提出了一种具有级联注意力解码的多尺度分层Transformer网络(MERIT),该网络在不同窗口大小下计算自注意力以捕捉有效的多尺度特征。
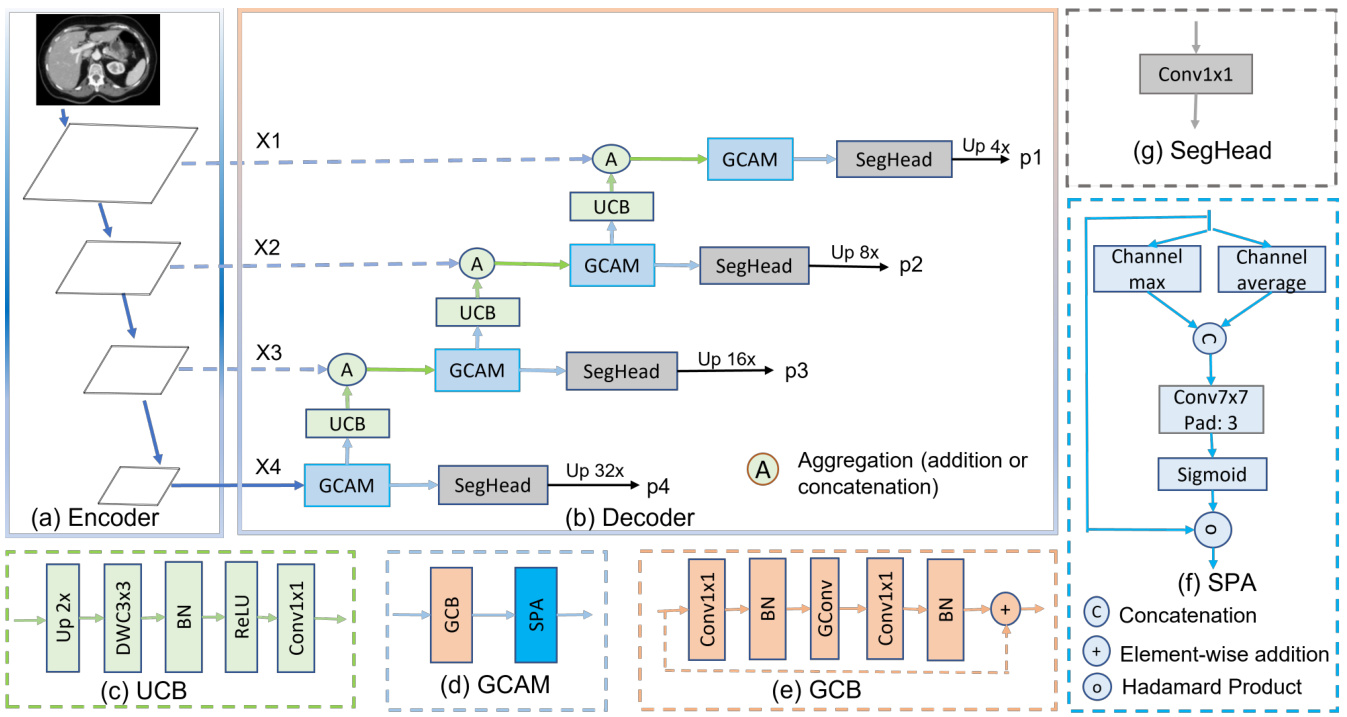
Figure 1. Hierarchical encoder with G-CASCADE network architecture. (a) PVTv2-b2 Encoder backbone with four stages, (b) GCASCADE decoder, (c) Up-convolution block (UCB), (d) Graph convolutional attention module (GCAM), (e) Graph convolution block (GCB), (f) Spatial attention (SPA), and (g) Segmentation head (SegHead). X1, X2, X3, and X4 are the output features of the four stages of hierarchical encoder. p1, p2, p3, and p4 are output segmentation maps from four stages of our decoder.
图 1: 采用 G-CASCADE 网络架构的层级编码器。(a) 包含四个阶段的 PVTv2-b2 编码器主干, (b) GCASCADE 解码器, (c) 上卷积块(UCB), (d) 图卷积注意力模块(GCAM), (e) 图卷积块(GCB), (f) 空间注意力(SPA), (g) 分割头(SegHead)。X1、X2、X3 和 X4 是层级编码器四个阶段的输出特征。p1、p2、p3 和 p4 是解码器四个阶段输出的分割图。
Attention mechanisms have also been explored in combination with both CNNs [27, 11] and transformer-based archi tec ture s [9] in medical image segmentation. PraNet [11] utilizes the reverse attention mechanism [6]. In PolypPVT [9], authors employ PVTv2 [40] as the encoder and integrates CBAM [43] attention blocks in the decoder, along with other modules. CASCADE [28] proposes a cascaded decoder that utilizes both channel attention [14] and spatial attention [5] modules for feature refinement. CASCADE extracts features from four stages of the transformer encoder and uses cascaded refinement to generate highresolution segmentation maps. Due to incorporating local information with global information of transformers, CASCADE exhibits remarkable performance in medical image segmentation. However, CASCADE decoder has two major limitations: this can lead to i) long-range attention deficit due using only convolution operations during decoding and ii) high computational inefficiency due to using three $3\times3$ convolutions in each stage of the decoder. We propose to use graph convolution to overcome these limitations.
注意力机制在医学图像分割领域已与CNN[27,11]和基于Transformer的架构[9]结合探索。PraNet[11]采用了反向注意力机制[6]。PolypPVT[9]使用PVTv2[40]作为编码器,并在解码器中整合了CBAM[43]注意力模块及其他组件。CASCADE[28]提出了一种级联解码器,同时利用通道注意力[14]和空间注意力[5]模块进行特征优化。该方法从Transformer编码器的四个阶段提取特征,通过级联优化生成高分辨率分割图。由于结合了Transformer的局部与全局信息,CASCADE在医学图像分割中表现出卓越性能。但该解码器存在两大缺陷:i)解码阶段仅使用卷积操作会导致长程注意力缺失;ii)解码器每阶段使用三个$3\times3$卷积导致计算效率低下。我们提出采用图卷积来克服这些局限。
3. Method
3. 方法
In this section, we first introduce a new G-CASCADE decoder, then explain two different transformer-based architectures (i.e., PVT-GCASCADE and MERITGCASCADE) incorporating our proposed decoder.
在本节中,我们首先介绍一种新的 G-CASCADE 解码器,然后阐述两种基于 Transformer 的不同架构 (即 PVT-GCASCADE 和 MERITGCASCADE) ,它们都整合了我们提出的解码器。
3.1. Cascaded Graph Convolutional Decoder (GCASCADE)
3.1. 级联图卷积解码器 (GCASCADE)
Existing transformer-based models have limited (local) contextual information processing ability among pixels. As a result, the transformer-based model faces difficulties in locating the more discriminating local features. To address this issue, some works [9, 28, 29] utilize computationally expensive 2D convolution blocks in the decoder. Although the convolution block helps to incorporate the local information, it results in long-range attention deficits. To overcome this problem, we propose a new cascaded graph convolutional decoder, G-CASCADE, for pyramid encoders.
现有基于Transformer的模型在像素间仅具备有限的(局部)上下文信息处理能力。这导致基于Transformer的模型难以定位更具区分性的局部特征。为解决该问题,部分研究[9,28,29]在解码器中采用了计算成本高昂的2D卷积模块。虽然卷积模块有助于整合局部信息,但会导致长程注意力缺失。为此,我们提出了一种面向金字塔编码器的新型级联图卷积解码器G-CASCADE。
As shown in Figure 1(b), G-CASCADE consists of efficient up-convolution blocks (UCBs) to upsample the features, graph convolutional attention modules (GCAMs) to robustly enhance the feature maps, and segmentation heads (SegHeads) to get the segmentation output. We have four GCAMs for the four stages of pyramid features from the encoder. To aggregate the multi-scale features, we first aggregate (e.g., addition or concatenation) the upsampled features from the previous decoder block with the features from the skip connections. Afterward, we process the concatenated features using our GCAM for enhancing semantic information. We then send the output from each GCAM to a prediction head. Finally, we aggregate four different prediction maps to produce the final segmentation output.
如图 1(b) 所示,G-CASCADE 由高效上卷积块 (UCB) 用于特征上采样、图卷积注意力模块 (GCAM) 用于鲁棒增强特征图,以及分割头 (SegHead) 用于生成分割输出。我们为编码器金字塔特征的四个阶段配置了四个 GCAM。为聚合多尺度特征,我们首先将前一解码器块的上采样特征与跳跃连接特征进行聚合 (如相加或拼接)。随后使用 GCAM 处理拼接后的特征以增强语义信息。每个 GCAM 的输出会被送入预测头,最终聚合四个不同预测图以生成最终分割结果。
3.1.1 Graph convolutional attention module (GCAM) 3.1.3 Segmentation head (SegHead)
3.1.1 图卷积注意力模块 (GCAM)
3.1.3 分割头 (SegHead)
We use the graph convolutional attention modules to refine the feature maps. GCAM consists of a graph convolution block $(G C B(.))$ to refine the features preserving long-range attention and a spatial attention [5] $(S P A(\cdot))$ block to capture the local contextual information as in Equation 1:
我们使用图卷积注意力模块来优化特征图。GCAM由一个图卷积块 $(G C B(.))$ 组成,用于通过保持长程注意力来优化特征,以及一个空间注意力 [5] $(S P A(\cdot))$ 块来捕获局部上下文信息,如公式1所示:
$$
G C A M(x)=S P A(G C B(x))
$$
$$
G C A M(x)=S P A(G C B(x))
$$
where $x$ is the input tensor and $G C A M(\cdot)$ represents the convolutional attention module. Due to using graph convolution, our GCAM is significantly more efficient than the convolutional attention module (CAM) proposed in [28].
其中 $x$ 是输入张量,$G C A M(\cdot)$ 表示卷积注意力模块。由于使用了图卷积,我们的 GCAM 比 [28] 中提出的卷积注意力模块 (CAM) 效率显著更高。
Graph Convolution Block (GCB): The GCB is used to enhance the features generated using our cascaded expanding path. In our GCB, we follow the Grapher design of Vision GNN [13]. GCB consists of a graph convolution layer $G C o n v(.)$ and two $1\times1$ convolution layers $C(\cdot)$ each followed by a batch normalization layer $B N(\cdot)$ and a ReLU activation layer $R(.).G C B(\cdot)$ is formulated as Equation 2:
图卷积块 (GCB): GCB用于增强通过级联扩展路径生成的特征。在我们的GCB中,我们遵循Vision GNN [13]的Grapher设计。GCB由一个图卷积层$G C o n v(.)$和两个$1\times1$卷积层$C(\cdot)$组成,每个卷积层后接批量归一化层$B N(\cdot)$和ReLU激活层$R(.)$。$G C B(\cdot)$的公式如式2所示:
$$
G C B(x)=R(B N(C(G C o n v(R(B N(C(x))))))
$$
$$
G C B(x)=R(B N(C(G C o n v(R(B N(C(x))))))
$$
where GConv can be formulated using Equation 3:
GConv 可以用公式 3 表示:
$$
G C o n v(x)=G E L U(B N(D y n C o n v(x)))
$$
$$
G C o n v(x)=G E L U(B N(D y n C o n v(x)))
$$
where DynConv(.) is a graph convolution (e.g., maxrelative, edge, GraphSAGE, and GIN) in dense dilated Knearest neighbour (KNN) graph. $B N(.)$ and $G E L U(.)$ are batch normalization and GELU activation, respectively.
其中DynConv(.)是密集扩张K近邻(KNN)图中的图卷积(例如maxrelative、edge、GraphSAGE和GIN)。$BN(.)$和$GELU(.)$分别表示批归一化和GELU激活函数。
SPatial Attention (SPA): The SPA determines where to focus in a feature map; then it enhances those features. The spatial attention is formulated as Equation 4:
空间注意力 (SPA): SPA决定在特征图中关注哪些位置,然后增强这些特征。空间注意力的计算公式如式4所示:
$$
S P A(x)=S i g m o i d(C o n v([C_{m a x}(x),C_{a v g}(x)]))\circledast x
$$
$$
S P A(x)=S i g m o i d(C o n v([C_{m a x}(x),C_{a v g}(x)]))\circledast x
$$
where Sigmoid(·) is a Sigmoid activation function. $C_{m a x}(\cdot)$ and $C_{a v g}(\cdot)$ represent the maximum and average values obtained along the channel dimension, respectively. $C o n v(\cdot)$ is a $7\times7$ convolution layer with padding 3 to enhance local contextual information (as in [9]). $\circledast$ is the Hadamard product.
其中Sigmoid(·)是Sigmoid激活函数。$C_{max}(\cdot)$和$C_{avg}(\cdot)$分别表示沿通道维度获取的最大值和平均值。$Conv(\cdot)$是一个填充为3的$7\times7$卷积层,用于增强局部上下文信息(如[9]所述)。$\circledast$表示哈达玛积。
3.1.2 Up-convolution block (UCB)
3.1.2 上卷积块 (UCB)
UCB progressively upsamples the features of the current layer to match the dimension to the next skip connection. Each UCB layer consists of an UpSampling $U p(\cdot)$ with scale-factor 2, a $3\times3$ depth-wise convolution $D W C(\cdot)$ with groups equal input channels, a batch normalization $B N(\cdot)$ , a $R e L U(.)$ activation, and a $1\times1$ convolution $C o n v(.)$ . The $U C B(\cdot)$ can be formulated as Equation 5:
UCB逐步对当前层的特征进行上采样,使其维度与下一个跳跃连接相匹配。每个UCB层包含一个缩放因子为2的上采样 $Up(\cdot)$、一个分组数等于输入通道数的 $3\times3$ 深度卷积 $DWC(\cdot)$、批归一化 $BN(\cdot)$、$ReLU(.)$ 激活函数以及一个 $1\times1$ 卷积 $Conv(.)$。$UCB(\cdot)$ 可表示为公式5:
$$
U C B(x)=C o n v(R e L U(B N(D W C(U p(x)))))
$$
$$
U C B(x)=C o n v(R e L U(B N(D W C(U p(x)))))
$$
Our UCB is light-weight as we replace the $3\times3$ convolution with a depth-wise convolution after upsampling.
我们的UCB设计轻量,因为在升采样后我们用深度卷积(depth-wise convolution)替代了$3\times3$卷积。
SegHead takes refined feature maps from the four stages of the decoder as input and predicts four output segmentation maps. Each SegHead layer consists of a $1\times1$ convolution $C o n v_{1\times1}(\cdot)$ which takes feature maps having $N_{i}$ channels ( $N_{i}$ is the number of channels in the feature map of stage $i\rrangle$ ) as input and gives output with channels equal to number of target classes for multi-class but 1 channel for binary prediction. The $S e g H e a d(\cdot)$ is formulated as Equation 6:
SegHead 以解码器四个阶段的精炼特征图作为输入,并预测四个输出分割图。每个 SegHead 层包含一个 $1\times1$ 卷积 $C o n v_{1\times1}(\cdot)$ ,该卷积以具有 $N_{i}$ 个通道的特征图 ( $N_{i}$ 是第 $i$ 阶段特征图的通道数 ) 作为输入,输出通道数等于多分类的目标类别数,但二分类预测时仅输出1个通道。 $S e g H e a d(\cdot)$ 的表达式如公式6所示:
$$
S e g H e a d(x)=C o n v_{1\times1}(x)
$$
$$
S e g H e a d(x)=C o n v_{1\times1}(x)
$$
3.2. Overall architecture
3.2. 整体架构
To ensure effective generalization and the ability to process multi-scale features in medical image segmentation, we integrate our proposed G-CASCADE decoder with two different hierarchical backbone encoder networks such as PVTv2 [40] and MERIT [29]. PVTv2 utilizes convolution operations instead of traditional transformer patch embedding modules to consistently capture spatial information. MERIT utilizes two MaxViT [34] encoders with varying window sizes for self-attention, thus enabling the capture of multi-scale features.
为确保医学图像分割中有效的泛化能力和多尺度特征处理能力,我们将提出的G-CASCADE解码器与两种不同的分层主干编码器网络(如PVTv2 [40]和MERIT [29])相结合。PVTv2采用卷积操作替代传统的Transformer补丁嵌入模块,以持续捕获空间信息。MERIT则利用两个具有不同窗口大小的MaxViT [34]编码器进行自注意力计算,从而实现对多尺度特征的捕捉。
By utilizing the PVTv2-b2 (Standard) encoder, we create the PVT-GCASCADE architecture. To adopt PVTv2- b2, we first extract the features (X1, X2, X3, and X4) from four layers and feed them (i.e., X4 in the upsample path and X3, X2, X1 in the skip connections) into our G-CASCADE decoder as shown in Figure 1(a-b). Then, the G-CASCADE processes them and produces four prediction maps that correspond to the four stages of the encoder network.
通过采用PVTv2-b2(标准)编码器,我们构建了PVT-GCASCADE架构。为适配PVTv2-b2,首先从四个层级提取特征(X1、X2、X3和X4),并将其(即上采样路径中的X4和跳跃连接中的X3、X2、X1)输入到G-CASCADE解码器,如图1(a-b)所示。随后,G-CASCADE处理这些特征并生成与编码器网络四个阶段相对应的四组预测图。
Besides, we introduce the new MERIT-GCASCADE architecture by adopting the architectural design of the MERIT network. In the case of MERIT, we only replace their decoder with our proposed decoder and keep their hybrid CNN-transformer MaxViT [34] encoder networks. In our MERIT-GCASCADE architecture, we extract hierarchical feature maps from four stages of first encoder and then feed them to the corresponding decoder. Afterwards, we aggregate the feedback from final stage of the decoder to the input image and feed them to second encoder having different window sizes for self-attention. We extract feature maps from four stages of the second decoder and feed them to the second decoder. We send cascaded skip connections like MERIT [29] to the second decoder. We get four output segmentation maps from the four stages of our second decoder. Finally, we aggregate the segmentation maps from the two decoders for four stages separately to produce four output segmentation maps. Our proposed decoder is designed to be adaptable and seamlessly integrates with other hierarchical backbone networks.
此外,我们通过采用MERIT网络的架构设计,引入了新的MERIT-GCASCADE架构。在MERIT的基础上,我们仅将其解码器替换为我们提出的解码器,并保留其混合CNN-Transformer MaxViT [34]编码器网络。在我们的MERIT-GCASCADE架构中,我们从第一个编码器的四个阶段提取分层特征图,然后将其馈送到相应的解码器。随后,我们将解码器最终阶段的反馈聚合到输入图像,并馈送到具有不同自注意力窗口大小的第二个编码器。我们从第二个解码器的四个阶段提取特征图并馈送到第二个解码器。与MERIT [29]类似,我们将级联跳跃连接发送到第二个解码器。从第二个解码器的四个阶段获得四个输出分割图。最后,我们分别聚合两个解码器四个阶段的分割图,生成四个输出分割图。我们提出的解码器设计具有适应性,并能与其他分层骨干网络无缝集成。
3.3. Multi-stage outputs and loss aggregation
3.3. 多阶段输出与损失聚合
We get four output segmentation maps $p_{1},p_{2},p_{3}$ , and $p_{4}$ from the four prediction heads for the four stages of our G-CASCADE decoder.
我们得到四个输出分割图 $p_{1},p_{2},p_{3}$ 和 $p_{4}$,分别对应G-CASCADE解码器四个阶段的预测头。
Output segmentation maps aggregation: We compute the final segmentation output using additive aggregation as in Equation 7:
输出分割图聚合:我们使用如公式7所示的加法聚合计算最终分割输出:
$$
s e g_o u t p u t=\alpha p_{1}+\beta p_{2}+\gamma p_{3}+\zeta p_{4}
$$
$$
s e g_o u t p u t=\alpha p_{1}+\beta p_{2}+\gamma p_{3}+\zeta p_{4}
$$
where $\alpha,\beta,\gamma$ , and $\zeta$ are the weights of each prediction head. We set $\alpha,\beta,\gamma$ , and $\zeta$ to 1.0 in all our experiments. We get the final prediction output by applying the Sigmoid activation for binary segmentation and Softmax activation for multi-class segmentation.
其中 $\alpha,\beta,\gamma$ 和 $\zeta$ 是每个预测头的权重。我们在所有实验中将 $\alpha,\beta,\gamma$ 和 $\zeta$ 设为1.0。对于二分类分割任务,我们使用Sigmoid激活函数得到最终预测输出;对于多分类分割任务,则使用Softmax激活函数。
Loss aggregation: Following MERIT [29], we use the combinatorial loss aggregation strategy, MUTATION in all our experiments. Therefore, we compute the loss for $2^{n}-1$ comb in a tro rial predictions synthesized from $n$ heads separately and then do a summation of them. We optimize this additive combinatorial loss during training.
损失聚合:遵循MERIT [29],我们在所有实验中使用组合损失聚合策略MUTATION。因此,我们分别计算由$n$个头合成的$2^{n}-1$种组合的损失,然后对它们求和。在训练过程中,我们优化这个加性组合损失。
4. Experimental Evaluation
4. 实验评估
In this section, we first describe the dataset and evaluation metrics followed by implementation details. Then, we conduct a comparative analysis between our proposed GCASCADE decoder-based architectures and SOTA methods to highlight the superior performance of our approach.
在本节中,我们首先描述数据集和评估指标,随后介绍实现细节。接着,通过对比分析我们提出的基于GCASCADE解码器的架构与SOTA方法,以凸显我们方法的优越性能。
4.1. Datasets
4.1. 数据集
We present the description of Synapse Multi-organ and ACDC datasets below. The description of ISIC2018, polyp, and retinal vessels segmentation datasets are available in supplementary materials (Section A).
我们在下文介绍Synapse多器官和ACDC数据集。ISIC2018、息肉及视网膜血管分割数据集的说明详见补充材料(章节A)。
Synapse Multi-organ dataset. The Synapse Multiorgan dataset1 contains 30 abdominal CT scans which have 3779 axial contrast-enhanced slices. Each CT scan has 85- 198 slices of $512\times512$ pixels. Similar to TransUNet [4], we divide the dataset randomly into 18 scans for training (2212 axial slices) and 12 scans for validation. We segment only 8 abdominal organs, i.e., aorta, gallbladder (GB), left kidney (KL), right kidney (KR), liver, pancreas (PC), spleen (SP), and stomach (SM).
Synapse多器官数据集。Synapse多器官数据集包含30个腹部CT扫描,共有3779张轴向增强对比切片。每个CT扫描包含85-198张$512\times512$像素的切片。与TransUNet [4]类似,我们将数据集随机划分为18个扫描用于训练(2212张轴向切片)和12个扫描用于验证。我们仅分割8个腹部器官,即主动脉、胆囊(GB)、左肾(KL)、右肾(KR)、肝脏、胰腺(PC)、脾脏(SP)和胃(SM)。
ACDC dataset. The ACDC dataset2 contains 100 cardiac MRI scans each of which consists of three organs, right ventricle (RV), myocardium (Myo), and left ventricle (LV). Following TransUNet [4], we use 70 cases (1930 axial slices) for training, 10 for validation, and 20 for testing.
ACDC数据集。ACDC数据集包含100例心脏MRI扫描,每例扫描包含三个器官:右心室(RV)、心肌(Myo)和左心室(LV)。按照TransUNet [4]的方法,我们使用70例(1930个轴向切片)进行训练,10例用于验证,20例用于测试。
4.2. Evaluation metrics
4.2. 评估指标
We use DICE, mIoU, and $95%$ Hausdorff Distance (HD95) to evaluate performance on the Synapse Multiorgan dataset. However, for the ACDC dataset, we use only DICE score as an evaluation metrics. We use DICE and mIoU as the evaluation metrics in polyp segmentation and ISIC2018 datasets. The DICE score $\bar{D}S\bar{C}(Y,\hat{Y})$ , $I o U(Y,{\hat{Y}})$ , and HD95 distance $D_{H}(Y,{\hat{Y}})$ are calculated using Equations 8, 9, and 10, respectively.
我们使用DICE、mIoU和95%豪斯多夫距离(HD95)来评估Synapse多器官数据集的性能。而对于ACDC数据集,我们仅采用DICE分数作为评估指标。在息肉分割和ISIC2018数据集中,我们使用DICE和mIoU作为评估指标。DICE分数$\bar{D}S\bar{C}(Y,\hat{Y})$、交并比$I o U(Y,{\hat{Y}})$以及HD95距离$D_{H}(Y,{\hat{Y}})$分别通过公式8、9和10计算得出。
$$
D S C(Y,{\hat{Y}})={\frac{2\times|Y\cap{\hat{Y}}|}{|Y|+|{\hat{Y}}|}}\times100\quad
$$
$$
D S C(Y,{\hat{Y}})={\frac{2\times|Y\cap{\hat{Y}}|}{|Y|+|{\hat{Y}}|}}\times100\quad
$$
$$
I o U(Y,\hat{Y})=\frac{|Y\cap\hat{Y}|}{|Y\cup\hat{Y}|}\times100
$$
$$
I o U(Y,\hat{Y})=\frac{|Y\cap\hat{Y}|}{|Y\cup\hat{Y}|}\times100
$$
where $Y$ and $\hat{Y}$ are the ground truth and predicted segmentation map, respectively.
其中 $Y$ 和 $\hat{Y}$ 分别表示真实分割图和预测分割图。
4.3. Implementation details
4.3. 实现细节
We use Pytorch 1.11.0 to implement our network and conduct experiments. We train all models on a single NVIDIA RTX A6000 GPU with 48GB of memory. We use the PVTv2-b2 and Small Cascaded MERIT as representative network. We use the pre-trained weights on ImageNet for both PVT and MERIT backbone networks. We train our model using AdamW optimizer [24] with both learning rate and weight decay of 0.0001.
我们使用Pytorch 1.11.0实现网络并进行实验。所有模型均在配备48GB内存的NVIDIA RTX A6000 GPU上进行训练。选用PVTv2-b2和Small Cascaded MERIT作为代表性网络,其骨干网络均采用ImageNet预训练权重。训练采用AdamW优化器[24],学习率和权重衰减率均设为0.0001。
GCB: We construct dense dilated graph using $K=11$ neighbors for KNN and use the Max-Relative (MR) graph convolution in all our experiments. The batch normalization is used after MR graph convolution. Following ViG [13], we also use the relative position vector for graph construction and reduction ratios of [1, 1, 4, 2] for graph convolution block in different stages.
GCB:我们使用 $K=11$ 个邻居构建密集扩张图进行KNN,并在所有实验中采用最大相对 (MR) 图卷积。MR图卷积后应用批量归一化。遵循ViG [13] 的方法,我们还利用相对位置向量进行图构建,并在不同阶段的图卷积块中采用 [1, 1, 4, 2] 的缩减比例。
Synapse Multi-organ dataset. We use a batch size of 6 and train each model for maximum of 300 epochs. We use the input resolution of $224\times224$ for PVT-GCASCADE and $256\times256$ , $224\times224;$ ) for MERIT-GCASCADE. We apply random rotation and flipping for data augmentation. The combined weighted Cross-entropy (0.3) and DICE (0.7) loss are utilized as the loss function.
Synapse多器官数据集。我们使用6的批量大小,每个模型最多训练300个周期。PVT-GCASCADE的输入分辨率为$224\times224$,MERIT-GCASCADE的输入分辨率为$256\times256$和$224\times224$。采用随机旋转和翻转进行数据增强。损失函数采用加权交叉熵(0.3)和DICE(0.7)的组合损失。
ACDC dataset. For the ACDC dataset, we train each model for a maximum of 150 epochs with a batch size of 12. We set the input resolution as $224\times224$ for PVTGCASCADE and $(256~\times256\$ , $224\times224_{,}$ ) for MERITGCASCADE. We apply random flipping and rotation for data augmentation. We optimize the combined weighted Cross-entropy (0.3) and DICE (0.7) loss function.
ACDC数据集。对于ACDC数据集,我们将每个模型的最大训练周期设为150,批次大小为12。PVTGCASCADE的输入分辨率设置为$224\times224$,MERITGCASCADE则设为$(256~\times256\$ , $224\times224_{,}$)。数据增强方面采用了随机翻转和旋转。优化目标为加权交叉熵(0.3)与DICE(0.7)的组合损失函数。
Table 1. Results of Synapse Multi-organ segmentation. We report only DICE scores for individual organs. We get the results of UNet, AttnUNet, PolypPVT, S S Former PV T, TransUNet, and SwinUNet from [28]. We reproduce the results of Cascaded MERIT with a batch size of 6. $\uparrow(\downarrow)$ denotes the higher (lower) the better. G-CASCADE results are averaged over five runs. The best results are shown in bold.
表 1: Synapse多器官分割结果。我们仅报告各器官的DICE分数。UNet、AttnUNet、PolypPVT、SSFormerPVT、TransUNet和SwinUNet的结果来自[28]。我们使用批大小为6复现了Cascaded MERIT的结果。$\uparrow(\downarrow)$ 表示越高(低)越好。G-CASCADE结果为五次运行的平均值,最佳结果以粗体显示。
| 架构 | DICE↑ | HD95↓ | mloU↑ | 主动脉 | 胆囊 | 左肾 | 右肾 | 肝脏 | 胰腺 | 脾脏 | 胃 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UNet [30] | 70.11 | 44.69 | 59.39 | 84.00 | 56.70 | 72.41 | 62.64 | 86.98 | 48.73 | 81.48 | 67.96 |
| AttnUNet [27] | 71.70 | 34.47 | 61.38 | 82.61 | 61.94 | 76.07 | 70.42 | 87.54 | 46.70 | 80.67 | 67.66 |
| R50+UNet [4] | 74.68 | 36.87 | 84.18 | 62.84 | 79.19 | 71.29 | 93.35 | 48.23 | 84.41 | 73.92 | |
| R50+AttnUNet [4] | 75.57 | 36.97 | 55.92 | 63.91 | 79.20 | 72.71 | 93.56 | 49.37 | 87.19 | 74.95 | |
| SSFormerPVT[38] | 78.01 | 25.72 | 67.23 | 82.78 | 63.74 | 80.72 | 78.11 | 93.53 | 61.53 | 87.07 | 76.61 |
| PolypPVT [9] | 78.08 | 25.61 | 67.43 | 82.34 | 66.14 | 81.21 | 73.78 | 94.37 | 59.34 | 88.05 | 79.40 |
| TransUNet [4] | 77.61 | 26.90 | 67.32 | 86.56 | 60.43 | 80.54 | 78.53 | 94.33 | 58.47 | 87.06 | 75.00 |
| SwinUNet [2] | 77.58 | 27.32 | 66.88 | 81.76 | 65.95 | 82.32 | 79.22 | 93.73 | 53.81 | 88.04 | 75.79 |
| MT-UNet [37] | 78.59 | 26.59 | 87.92 | 64.99 | 81.47 | 77.29 | 93.06 | 59.46 | 87.75 | 76.81 | |
| MISSFormer [16] | 81.96 | 18.20 | 86.99 | 68.65 | 85.21 | 82.00 | 94.41 | 65.67 | 91.92 | 80.81 | |
| PVT-CASCADE [28] | 81.06 | 20.23 | 70.88 | 83.01 | 70.59 | 82.23 | 80.37 | 94.08 | 64.43 | 90.10 | 83.69 |
| TransCASCADE [28] | 82.68 | 17.34 | 73.48 | 86.63 | 68.48 | 87.66 | 84.56 | 94.43 | 65.33 | 90.79 | 83.52 |
| Cascaded MERIT [29] | 84.32 | 14.27 | 75.44 | 86.67 | 72.63 | 87.71 | 84.62 | 95.02 | 70.74 | 91.98 | 85.17 |
| PVT-GCASCADE (Ours) | 83.28 | 15.83 | 73.91 | 86.50 | 71.71 | 87.07 | 83.77 | 95.31 | 66.72 | 90.84 | 83.58 |
| MERIT-GCASCADE (Ours) | 84.54 | 10.38 | 75.83 | 88.05 | 74.81 | 88.01 | 84.83 | 95.38 | 69.73 | 91.92 | 83.63 |
Table 2. Results on ACDC dataset. DICE scores are reported for individual organs. We get the results of SwinUNet from [28]. GCASCADE results are averaged over five runs. The best results are shown in bold.
表 2: ACDC数据集上的结果。DICE分数针对各个器官单独报告。我们从[28]中获取了SwinUNet的结果。GCASCADE结果是五次运行的平均值。最佳结果以粗体显示。
| 方法 | 平均 | Dice | RV | Myo | LV |
|---|---|---|---|---|---|
| R50+UNet[4] | 87.55 | 87.10 | 80.63 | 94.92 | |
| R50+AttnUNet[4] | 86.75 | 87.58 | 79.20 | 93.47 | |
| ViT+CUP [4] | 81.45 | 81.46 | 70.71 | 92.18 | |
| R50+ViT+CUP [4] | 87.57 | 86.07 | 81.88 | 94.75 | |
| TransUNet [4] | 89.71 | 86.67 | 87.27 | 95.18 | |
| SwinUNet [2] | 88.07 | 85.77 | 84.42 | 94.03 | |
| MT-UNet [37] | 90.43 | 86.64 | 89.04 | 95.62 | |
| MISSFormer [16] | 90.86 | 89.55 | 88.04 | 94.99 | |
| PVT-CASCADE[28] | 91.46 | 89.97 | 88.9 | 95.50 | |
| TransCASCADE [28] | 91.63 | 90.25 | 89.14 | 95.50 | |
| Cascaded MERIT [29] | 91.85 | 90.23 | 89.53 | 95.80 | |
| PVT-GCASCADE (Ours) | 91.95 | 90.31 | 89.63 | 95.91 | |
| MERIT-GCASCADE (Ours) | 92.23 | 90.64 | 89.96 | 96.08 |
ISIC2018 dataset: We resize the images into $384\times384$ resolution. Then, we train our model for 200 epochs with a batch size of 4 and a gradient clip of 0.5. We optimize the combined weighted BCE and weighted IoU loss function.
ISIC2018数据集:我们将图像分辨率调整为$384\times384$,随后以批量大小4和梯度裁剪值0.5训练模型200轮次,优化组合加权BCE与加权IoU损失函数。
Polyp datasets. We resize the image to $352\times352$ and use a multi-scale ${0.75,1.0,1.25}$ training strategy with a gradient clip limit of 0.5 like CASCADE [28]. We use a batch size of 4 and train each model a maximum of 200 epochs. We optimize the combined weighted BCE and weighted IoU loss function.
息肉数据集。我们将图像大小调整为$352\times352$,并采用类似CASCADE [28]的多尺度${0.75,1.0,1.25}$训练策略,梯度裁剪限制为0.5。使用批量大小为4,每个模型最多训练200个周期。我们优化了加权BCE和加权IoU的组合损失函数。
4.4. Results
4.4. 结果
We compare our architectures (i.e., PVT-GCASCADE and MERIT-GCASCADE) with SOTA CNN and transformer-based segmentation methods on Synapse Multi-organ, ACDC, ISIC2018 [8], and Polyp (i.e., Endoscene [35], CVC-ClinicDB [1], Kvasir [18], ColonDB [32]) datasets. The results of ISIC2018, polyp, and retinal vessels segmentation datasets are reported in the supplementary materials (Section B).
我们在Synapse多器官、ACDC、ISIC2018 [8]和息肉(即Endoscene [35]、CVC-ClinicDB [1]、Kvasir [18]、ColonDB [32])数据集上,将我们的架构(即PVT-GCASCADE和MERIT-GCASCADE)与基于SOTA CNN和Transformer的分割方法进行了比较。ISIC2018、息肉和视网膜血管分割数据集的结果见补充材料(B节)。
4.4.1 Quantitative results on Synapse Multi-organ dataset
4.4.1 Synapse多器官数据集的定量结果
Table 1 presents the performance of different CNNand transformer-based methods on Synapse Multi-organ segmentation dataset. We can see from Table 1 that our MERIT-GCASCADE significantly outperforms all the SOTA CNN- and transformer-based 2D medical image segmentation methods thus achieving the best average DICE score of $84.54%$ . Our PVT-GCASCADE and MERIT-GCASCADE outperforms their counterparts PVTCASCADE and Cascaded MERIT by $2.22%$ and $0.22%$ DICE scores, respectively with significantly lower computational costs. Similarly, our PVT-GCASCADE and MERIT-GCASCADE outperforms their counterparts by 4.4 and 3.89 in HD95 distance. Our MERIT-GCASCADE has the lowest HD95 distance (10.38) which is 3.89 lower than the best SOTA method Cascaded MERIT (HD95 of 14.27). The lower HD95 scores indicate that our G-CASCADE decoder can better locate the boundary of organs.
表 1: 展示了不同基于CNN和Transformer的方法在Synapse多器官分割数据集上的性能。从表1可以看出,我们的MERIT-GCASCADE显著优于所有基于CNN和Transformer的2D医学图像分割SOTA方法,从而实现了最佳平均DICE分数 $84.54%$ 。我们的PVT-GCASCADE和MERIT-GCASCADE分别以 $2.22%$ 和 $0.22%$ 的DICE分数优势超越对应方法PVTCASCADE和Cascaded MERIT,同时计算成本显著降低。类似地,我们的PVT-GCASCADE和MERIT-GCASCADE在HD95距离上分别以4.4和3.89的优势超越对应方法。我们的MERIT-GCASCADE具有最低的HD95距离(10.38),比最佳SOTA方法Cascaded MERIT(HD95为14.27)低3.89。较低的HD95分数表明我们的G-CASCADE解码器能更好地定位器官边界。
Our proposed decoder also shows boost in the DICE scores of individual organ segmentation. We can see from the Table 1 that our proposed MERIT-GCASCADE significantly outperforms SOTA methods on five out of eight organs. We believe that G-CASCADE decoder demonstrates better performance due to using graph convolution together with the transformer encoder.
我们提出的解码器在单个器官分割的DICE分数上也显示出提升。从表1可以看出,我们提出的MERIT-GCASCADE在八个器官中的五个上显著优于SOTA方法。我们认为G-CASCADE解码器由于结合了图卷积和Transformer编码器,因此表现出更好的性能。
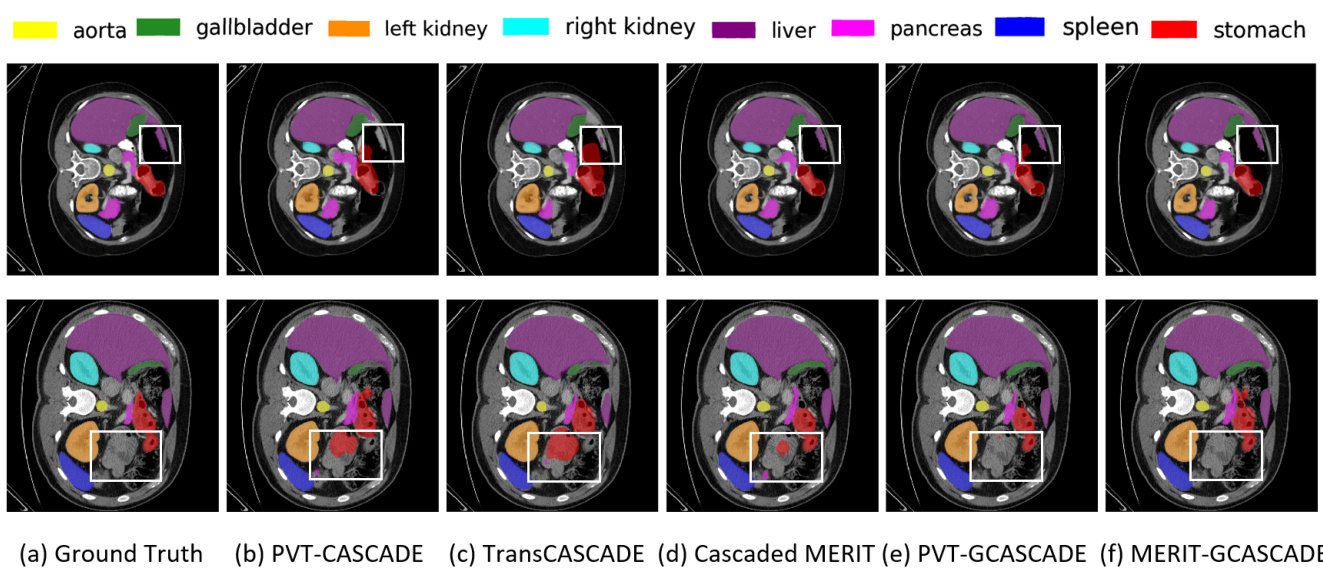
Figure 2. Qualitative results on Synapse multi-organ dataset. (a) Ground Truth (GT), (b) PVT-CASCADE, (c) Trans CASCADE, (d) Cascaded MERIT, (e) PVT-GCASCADE, and (f) MERIT-GCASCADE. We overlay the segmentation maps on top of original image/slice. We use the white bounding box to highlight regions where most of the methods have incorrect predictions.
图 2: Synapse多器官数据集的定性分析结果。(a) 真实标注 (GT), (b) PVT-CASCADE, (c) Trans CASCADE, (d) Cascaded MERIT, (e) PVT-GCASCADE, (f) MERIT-GCASCADE。我们在原始图像/切片上叠加了分割结果图,并用白色边框标明了多数方法预测错误的区域。
4.4.2 Quantitative results on ACDC dataset
4.4.2 ACDC数据集上的定量结果
We have conducted another set of experiments on the MRI images of the ACDC dataset using our architectures. Table 2 presents the average DICE scores of our PVTGCASCADE and MERIT-GCASCADE along with other SOTA methods. Our MERIT-GCASCADE achieves the highest average DICE score of $92.23%$ thus improving about $0.38%$ over Cascaded MERIT though our decoder has significantly lower computational cost (see Table 5). Our PVT-GCASCADE gains $91.95%$ DICE score which is also better than all other methods. Besides, both our PVTGCASCADE and MERIT-GCASCADE have better DICE scores in all three organs segmentation.
我们在ACDC数据集的MRI图像上使用我们的架构进行了另一组实验。表2展示了我们的PVTGCASCADE和MERIT-GCASCADE以及其他SOTA方法的平均DICE分数。我们的MERIT-GCASCADE实现了最高的平均DICE分数$92.23%$,比Cascaded MERIT提高了约$0.38%$,尽管我们的解码器计算成本显著更低(见表5)。我们的PVT-GCASCADE获得了$91.95%$的DICE分数,也优于所有其他方法。此外,我们的PVTGCASCADE和MERIT-GCASCADE在所有三个器官分割中都取得了更好的DICE分数。
4.4.3 Qualitative results on Synapse Multi-organ dataset
4.4.3 Synapse多器官数据集的定性结果
We present the segmentation outputs of our proposed method and three other SOTA methods on two sample images in Figure 2. If we look into the highlighted regions in both samples, we can see that MERIT-GCASCADE consistently segments the organs with minimal false negative and false positive results. PVT-GCASCADE and Cascaded MERIT show comparable results. PVT-GCASCADE has false positives in first sample (i.e., first row) and has better segmentat
