PMC-VQA: Visual Instruction Tuning for Medical Visual Question Answering
PMC-VQA:面向医疗视觉问答的视觉指令微调
Xiaoman Zhang $^{* ,1,2}$ , Chaoyi Wu $^{* ,1,2}$ , Ziheng Zhao $^{1,2}$ , Weixiong Lin $^{1,2}$ , Ya Zhang $^{1,2}$ , Yanfeng Wang $^{1,2,}$ ,† and Weidi Xie $^{1,2}$ ,† $^{1}$ Shanghai Jiao Tong University Shanghai AI Laboratory
Xiaoman Zhang$^{* ,1,2}$, Chaoyi Wu$^{* ,1,2}$, Ziheng Zhao$^{1,2}$, Weixiong Lin$^{1,2}$, Ya Zhang$^{1,2}$, Yanfeng Wang$^{1,2,}$†, Weidi Xie$^{1,2}$†
$^{1}$上海交通大学
$^{2}$上海人工智能实验室
Medical Visual Question Answering (MedVQA) presents a significant opportunity to enhance diagnostic accuracy and healthcare delivery by leveraging artificial intelligence to interpret and answer questions based on medical images. In this study, we reframe the problem of MedVQA as a generation task that naturally follows the human-machine interaction and propose a generative-based model for medical visual understanding by aligning visual information from a pre-trained vision encoder with a large language model. We establish a scalable pipeline to construct a large-scale medical visual question-answering dataset, named PMC-VQA, which contains 227k VQA pairs of 149k images that cover various modalities or diseases. We train the proposed model on PMC-VQA and then fine-tune it on multiple public benchmarks, e.g., VQA-RAD, SLAKE, and Image-Clef-2019, significantly outperforming existing MedVQA models in generating relevant, accurate free-form answers. In addition, we propose a test set that has undergone manual verification, which is significantly more challenging, serving to better monitor the development of generative MedVQA methods. To facilitate comprehensive evaluation and comparison, we have maintained a leader board at https://papers with code.com/paper/pmc-vqa-visual-instruction-tuning-for-medical, offering a centralized resource for tracking progress and benchmarking state-of-the-art approaches. The PMC-VQA dataset emerges as a vital resource for the field of research, and the MedVInT presents a significant breakthrough in the area of MedVQA.
医学视觉问答 (MedVQA) 通过利用人工智能 (AI) 解释和回答基于医学图像的问题,为提高诊断准确性和医疗服务提供了重要机遇。本研究将 MedVQA 问题重新定义为一种自然遵循人机交互的生成任务,并提出了一种基于生成的医学视觉理解模型,通过将预训练视觉编码器的视觉信息与大语言模型对齐来实现。我们建立了一个可扩展的流程来构建名为 PMC-VQA 的大规模医学视觉问答数据集,该数据集包含 149k 张图像的 227k 个 VQA 对,涵盖多种模态或疾病。我们在 PMC-VQA 上训练所提出的模型,然后在多个公共基准 (如 VQA-RAD、SLAKE 和 Image-Clef-2019) 上进行微调,在生成相关、准确的自由形式答案方面显著优于现有的 MedVQA 模型。此外,我们提出了一个经过人工验证的测试集,该测试集更具挑战性,有助于更好地监测生成式 MedVQA 方法的发展。为了促进全面评估和比较,我们在 https://paperswithcode.com/paper/pmc-vqa-visual-instruction-tuning-for-medical 上维护了一个排行榜,为跟踪进展和基准测试最先进方法提供了集中资源。PMC-VQA 数据集成为该研究领域的重要资源,而 MedVInT 在 MedVQA 领域取得了重大突破。
1 Introduction
1 引言
Large language models (LLMs), such as GPT-4 [43], Med-PaLM [51], PMC-LLaMA [57] have recently achieved remarkable success in the field of medical natural language processing [24, 27, 42]. While recent LLMs excel in language understanding in the medical domain, they are essentially “blind” to visual modalities, such as images and videos, hindering the use of visual content as inputs. This limitation is particularly evident in the Medical Visual Question Answering (MedVQA) domain, where there is a critical need for models to interpret medical visual content to answer text-based queries accurately [33].
大语言模型 (LLMs) ,例如 GPT-4 [43] 、Med-PaLM [51] 、PMC-LLaMA [57] 最近在医学自然语言处理领域取得了显著成功 [24, 27, 42] 。尽管当前的大语言模型在医学领域的语言理解方面表现出色,但它们本质上对视觉模态 (例如图像和视频) 是"盲视"的,这阻碍了将视觉内容作为输入的使用。这一局限性在医学视觉问答 (MedVQA) 领域尤为明显,该领域迫切需要模型能够解读医学视觉内容以准确回答基于文本的查询 [33] 。
MedVQA is an important and emerging field at the intersection of artificial intelligence and healthcare, which involves developing systems that can understand and interpret medical images and provide relevant answers to questions posed about these images. By integrating AI with medical expertise, MedVQA aims to significantly impact healthcare outcomes, patient care, and medical science [60, 53]. For example, the MedVQA system can enhance diagnostic accuracy for clinicians, improve patient understanding of medical information, and advance medical education and research.
MedVQA是人工智能与医疗交叉领域中一个重要且新兴的方向,其核心是开发能够理解医学影像并针对影像相关问题提供答案的系统。通过将AI与医学专业知识相结合,MedVQA致力于对医疗结果、患者护理和医学研究产生深远影响 [60, 53]。例如,该系统可提升临床医生的诊断准确率,改善患者对医学信息的理解,并推动医学教育与研究发展。
However, existing MedVQA methods [40, 34, 11, 32] typically treat the problem as a retrieval task with a limited answer base and train multi-modal vision-language models with contrastive or classification objectives. Consequently, they are only useful for limited use cases where a finite set of outcomes is provided beforehand. We propose to develop the first open-ended MedVQA system with a generative model as the backend, capable of handling diverse questions that arise in clinical practice, generating answers in free form without being constrained by the vocabulary. While there has been promising research in visual-language representation learning, such as Flamingo [1] and BLIP [30], these models have primarily been trained on natural language and images, with very limited application in the medical domain, due to the complex and nuanced visual concepts often found in medical scenarios.
然而,现有的医学视觉问答 (MedVQA) 方法 [40, 34, 11, 32] 通常将该问题视为答案库有限的检索任务,并通过对比或分类目标训练多模态视觉-语言模型。因此,这些方法仅适用于预先提供有限结果集的特定场景。我们提出开发首个基于生成式模型 (generative model) 的后端开放式 MedVQA 系统,能够处理临床实践中出现的多样化问题,并以自由形式生成答案,不受预定义词汇表的限制。尽管视觉-语言表征学习领域已取得突破性进展(例如 Flamingo [1] 和 BLIP [30]),但这些模型主要基于自然语言和普通图像进行训练,由于医学场景中复杂而细微的视觉概念,其在医疗领域的应用仍非常有限。
To effectively train the generative-based models, our study reveals that existing datasets are limited in size, making them insufficient for training high-performing models. we leverage well-established medical visual-language datasets [32] and initiate a scalable, automatic pipeline for constructing a new large-scale medical visual question-answering dataset. This new dataset, termed as PMC-VQA, contains 227k VQA pairs of 149k images, including 80% of radiological images, covering various modalities or diseases (Figure 1), surpassing existing datasets in terms of both amount and diversity.
为有效训练基于生成式 AI (Generative AI) 的模型,我们的研究表明现有数据集规模有限,难以支撑高性能模型的训练。为此,我们利用成熟的医学视觉语言数据集[32],构建了一个可扩展的自动化流程来创建新的大规模医学视觉问答数据集。这个名为 PMC-VQA 的新数据集包含 14.9 万张图像对应的 22.7 万个视觉问答对,其中 80% 为放射影像,涵盖多种模态或疾病 (图 1),在数据量和多样性方面均超越现有数据集。
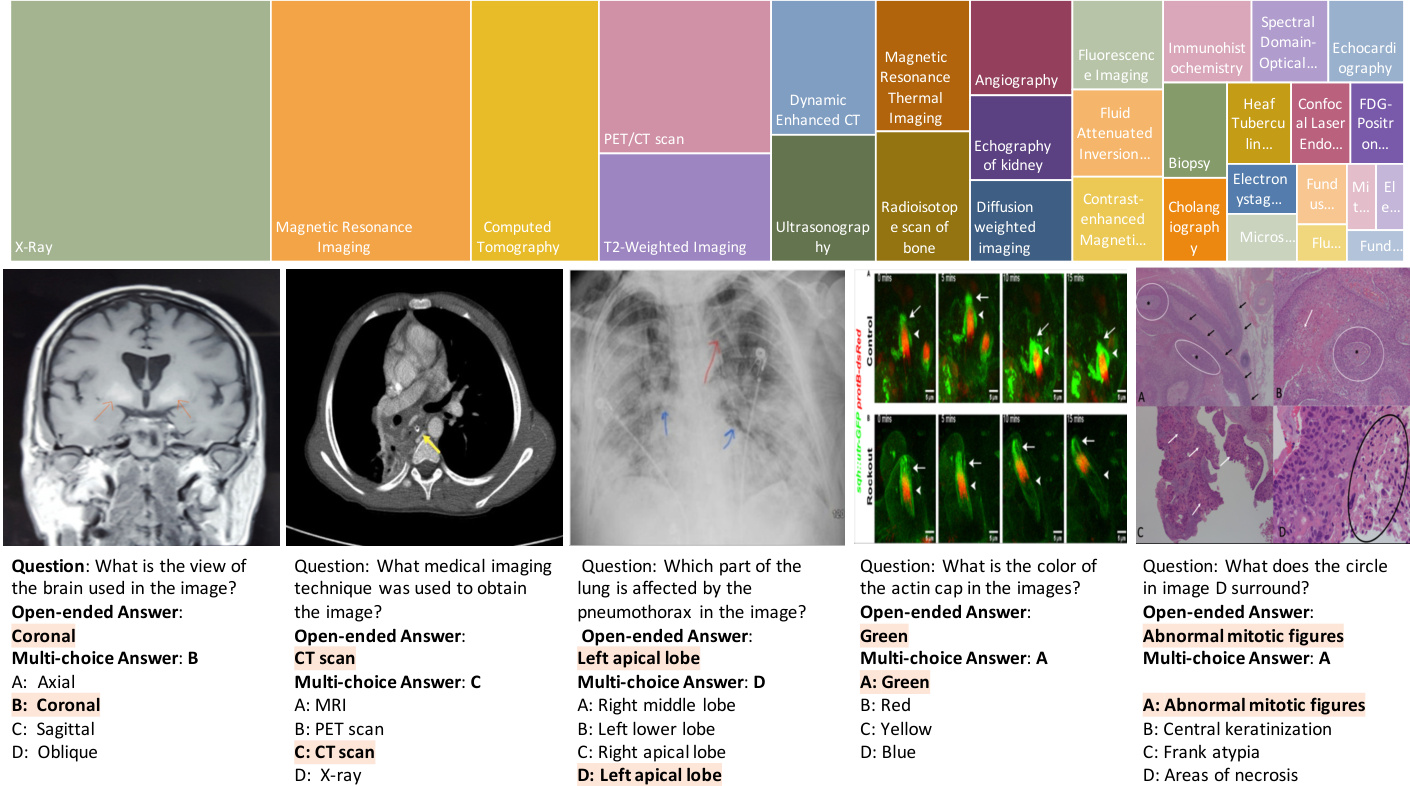
Figure 1 | (a) Several examples of challenging questions and answers along with their respective images. To answer questions related to these images, the network must acquire sufficient medical knowledge, for example, for the first two images, it is essential to recognize the anatomy structure and modalities; for the third image, recognizing the X-ray image pattern of path o logie s is necessary; for the final two images, apart from the basic biomedical knowledge, the model is also required to discern colors, differentiate subfigures, and perform Optical Character Recognition (OCR). (b) The top 20 figure types in PMC-VQA, cover a wide range of diagnostic procedures.
图 1 | (a) 具有挑战性的问题与答案示例及其对应图像。要回答与这些图像相关的问题,网络必须掌握足够的医学知识,例如:对于前两张图像,识别解剖结构和成像模态至关重要;对于第三张图像,需识别病理特征的X光影像模式;最后两幅图像则要求模型除基础生物医学知识外,还需辨别颜色、区分子图并进行光学字符识别(OCR)。(b) PMC-VQA数据集中排名前20的图表类型,涵盖了广泛的诊断流程。
In our experiments, we trained a generative visual-language model, termed as MedVInT, on the training set of PMC-VQA and fine-tuned it on the existing public benchmarks, e.g., VQA-RAD [28], SLAKE [35], and ImageClef-VQA-2019 [6]. outperforming existing models by a large margin, achieving over 80% accuracy on multi-choice selection. However, while evaluating our proposed challenging benchmark, even the state-of-the-art models struggle, showing that there is still ample room for development in this field.
在我们的实验中,我们在PMC-VQA训练集上训练了一个生成式视觉语言模型(MedVInT),并在现有公开基准(如VQA-RAD [28]、SLAKE [35]和ImageClef-VQA-2019 [6])上进行了微调。该模型以显著优势超越现有模型,在多选题任务中准确率超过80%。然而,在评估我们提出的挑战性基准时,即使最先进的模型也表现不佳,这表明该领域仍有广阔发展空间。
In summary, our contributions are as follows: (i) We reframe the problem of MedVQA as a generative learning task and propose MedVInT, a model obtained by aligning a pre-trained vision encoder with a large language model through visual instruction tuning; (ii) We introduce a scalable pipeline and construct a large-scale MedVQA dataset, PMC-VQA, which far exceeds the size and diversity of existing datasets, covering various modalities and diseases; (iii) We pre-train MedVInT on PMC-VQA and fine-tune it on VQA-RAD [28] and SLAKE [35], achieving state-of-the-art performance and significantly outperforming existing models; (iv) We propose a new test set and present a more challenging benchmark for MedVQA, to evaluate the performance of VQA methods thoroughly.
总之,我们的贡献如下:(i) 我们将医学视觉问答 (MedVQA) 问题重新定义为生成式学习任务,并提出通过视觉指令微调将预训练视觉编码器与大语言模型对齐的 MedVInT 模型;(ii) 我们引入可扩展的流程并构建了大规模 MedVQA 数据集 PMC-VQA,其规模和多样性远超现有数据集,涵盖多种模态和疾病;(iii) 我们在 PMC-VQA 上预训练 MedVInT,并在 VQA-RAD [28] 和 SLAKE [35] 上微调,实现了最先进的性能,显著优于现有模型;(iv) 我们提出了新的测试集,为 MedVQA 建立了更具挑战性的基准,以全面评估 VQA 方法的性能。
2 Results
2 结果
The goal of our proposed model, Medical Visual Instruction Tuning (MedVInT), is to perform generative-based medical visual question answering (MedVQA). Serving for this purpose, we curate a new large-scale medical visual instruction tuning dataset, namely PMC-VQA. In this section, we start by a comprehensive analysis on the PMC-VQA dataset, which contains 227k VQA pairs of 149k images, covering various modalities or diseases and compare it with the existing medical VQA datasts. Then, we will evaluate our trained model on three external MedVQA benchmarks, VQA-RAD [28], SLAKE [35] and ImageClef-VQA-2019 [6]. Note that, our model has two variants, which are tailored to encoder-based and decoder-based language models, respectively, denoted as MedVInT-TE and MedVInT-TD. At last, we establish a novel generative MedVQA benchmark with PMC-VQA, and evaluate various pre-trained visual or language models using our framework, serving as a reference to promote the future research in generative medical VQA.
我们提出的模型 Medical Visual Instruction Tuning (MedVInT) 的目标是基于生成式方法执行医学视觉问答 (MedVQA)。为此,我们构建了一个新的大规模医学视觉指令微调数据集 PMC-VQA。本节首先对 PMC-VQA 数据集进行全面分析,该数据集包含 149k 张图像的 227k 个 VQA 对,涵盖多种模态或疾病,并与现有医学 VQA 数据集进行对比。随后,我们将在三个外部 MedVQA 基准测试 (VQA-RAD [28]、SLAKE [35] 和 ImageClef-VQA-2019 [6]) 上评估训练好的模型。需注意的是,我们的模型有两个变体,分别针对基于编码器和基于解码器的大语言模型定制,记为 MedVInT-TE 和 MedVInT-TD。最后,我们基于 PMC-VQA 建立了一个新的生成式 MedVQA 基准,并使用我们的框架评估了各种预训练视觉或语言模型,为促进生成式医学 VQA 的未来研究提供参考。
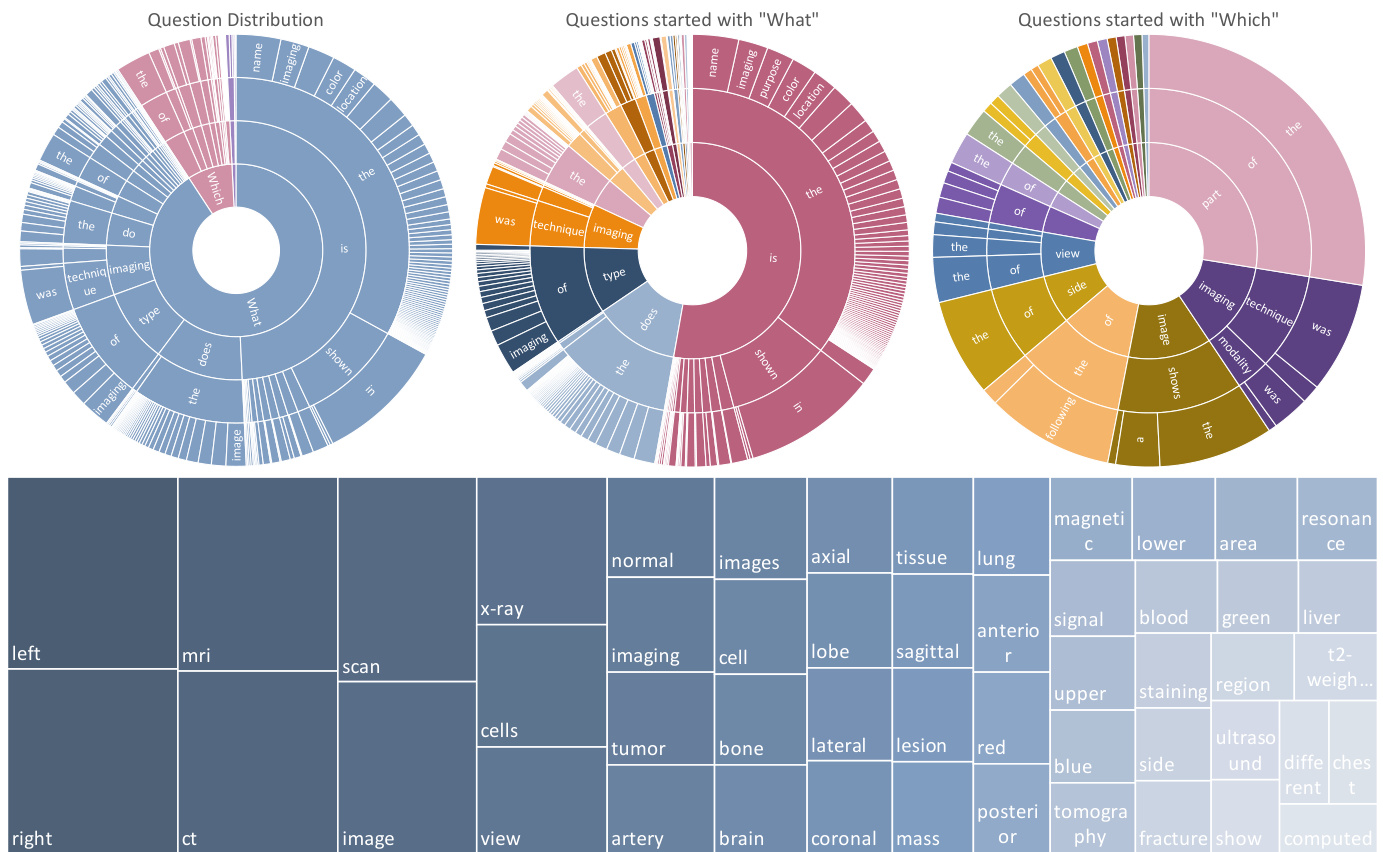
Figure 2 | The top row shows the question distribution of the training set by their first four words. From left to right are all questions, questions started with “What” and questions started with “Which”. The ordering of the words starts towards the center and radiates outwards. The bottom row show the answer distribution of the training set.
图 2 | 首行展示了训练集中问题按其前四个词的分布情况。从左至右分别为:全部问题、以"What"开头的问题、以"Which"开头的问题。词汇排序从中心向外辐射排列。次行显示训练集的答案分布情况。
2.1 Data Analysis
2.1 数据分析
This section provides an analysis of images, questions, and answers of our final proposed dataset. In detail, the dataset comprises 227k image-question pairs, some examples are presented in Figure 1, which demonstrates the wide diversity of images within our dataset. As indicated in Table 1, PMC-VQA outperforms existing MedVQA datasets in terms of data size and modality diversity. The questions in our dataset cover a range of difficulties, from simple questions such as identifying image modalities, perspectives, and organs to challenging questions that require specialized knowledge and judgment. Additionally, our dataset includes difficult questions that demand the ability to identify the specific target sub-figure from the compound figure.
本节对我们最终提出的数据集中的图像、问题和答案进行了分析。具体而言,该数据集包含227k个图像-问题对,图1展示了一些示例,体现了数据集中图像的广泛多样性。如表1所示,PMC-VQA在数据规模和模态多样性方面优于现有的MedVQA数据集。我们数据集中的问题涵盖了一系列难度,从识别图像模态、视角和器官等简单问题,到需要专业知识和判断的挑战性问题。此外,我们的数据集还包括需要从复合图中识别特定目标子图的难题。
Our analysis of the PMC-VQA dataset can be summarized in three aspects: (i) Images: We show the top 20 figure types in Figure 1. The images in the PMC-VQA are extremely diverse, ranging from Radiology to Signals. (ii) Questions: We clustered the questions into different types based on the words that start the question, as shown in Figure 2. The dataset covers very diverse question types, including “What is the difference...”, “What type of imaging...”, and “Which image shows...”. Most questions range from 5 to 15 words, and detailed information about the distribution of question lengths is shown in the supplementary materials A.1. (iii) Answers: The words in answers primarily encompass positional descriptions, image modalities, and specific anatomical regions. Detailed information about the top 50 words that appeared in the
我们对PMC-VQA数据集的分析可总结为三个方面:(i) 图像:图1展示了前20种图像类型。PMC-VQA中的图像极其多样,涵盖从放射学到信号处理等领域。(ii) 问题:我们根据问题起始词对问题进行了分类,如图2所示。该数据集覆盖了多样化的问题类型,包括"差异是什么..."、"哪种成像类型..."以及"哪张图像显示..."等。大多数问题长度在5到15个单词之间,问题长度分布的详细信息见补充材料A.1。(iii) 答案:答案中的词汇主要包含位置描述、图像模态和特定解剖区域。关于答案中出现频率最高的50个词汇的详细信息...
Table 1 | Comparison of existing medical VQA datasets with PMC-VQA, demonstrating our dataset’s significant increase in size and diversity. Mixture refers to Radiology, Pathology, Microscopy, Signals, Generic biomedical illustrations, etc.
| Dataset | Modality | Source | Images | QA1 pairs |
| VQA-RAD [28] | Radiology | MedPix? database | 0.3k | 3.5k |
| PathVQA [22] | Pathology | PEIR Digital Library [25] | 5k | 32.8k |
| SLAKE [35] | Radiology | MSD [3], ChestX-ray8 [56], CHAOS [26] | 0.7k | 14k |
| VQA-Med-2021 | Radiology | MedPix database | 5k | 5k |
| PMC-VQA | Mixture* (80% Radiology) | PubMed Central? | 149k | 227k |
表 1 | 现有医学VQA数据集与PMC-VQA的对比,展示本数据集在规模和多样性上的显著提升。Mixture指放射学、病理学、显微镜、信号、通用生物医学插图等。
| 数据集 | 模态 | 来源 | 图像 | 问答对 |
|---|---|---|---|---|
| VQA-RAD [28] | 放射学 | MedPix数据库 | 0.3k | 3.5k |
| PathVQA [22] | 病理学 | PEIR数字图书馆 [25] | 5k | 32.8k |
| SLAKE [35] | 放射学 | MSD [3], ChestX-ray8 [56], CHAOS [26] | 0.7k | 14k |
| VQA-Med-2021 | 放射学 | MedPix数据库 | 5k | 5k |
| PMC-VQA | Mixture* (80%放射学) | PubMed Central | 149k | 227k |
2.2 Evaluation on Public Benchmarks
2.2 公开基准测试评估
Table 2 presents the performance of our MedVInT model on three widely recognized MedVQA benchmarks: VQA-RAD, SLAKE, and ImageClef-VQA-2019. The results demonstrate that the MedVInT model, regardless of whether we use the “MedVInT-TE” or “MedVInT-TD” version, surpasses previous best-performing methods on the VQA-RAD and SLAKE datasets. By default, we employ PMC-CLIP as the visual backbone and PMC-LLaMA as the language backbone, as demonstrated in Table 3, models pre-trained using PubMed Central data generally yield superior performance.
表 2 展示了我们的 MedVInT 模型在三个广泛认可的 MedVQA 基准测试 (VQA-RAD、SLAKE 和 ImageClef-VQA-2019) 上的性能表现。结果表明,无论是使用 "MedVInT-TE" 还是 "MedVInT-TD" 版本,MedVInT 模型在 VQA-RAD 和 SLAKE 数据集上都超越了先前的最佳方法。默认情况下,我们采用 PMC-CLIP 作为视觉主干网络,PMC-LLaMA 作为语言主干网络。如表 3 所示,使用 PubMed Central 数据预训练的模型通常能获得更优的性能。
It is important to note that both the VQA-RAD and SLAKE datasets include questions that are categorized as either open-ended or close-ended. Close-ended questions restrict answers to a predefined set of options, whereas open-ended questions allow for free-from text responses. Specifically, for open-ended questions, the accuracy rates were enhanced from $67.2%$ to $73.7%$ on VQA-RAD and from $81.9%$ to $88.2%$ on SLAKE. For close-ended questions, the MedVInT model improved the accuracy from $84.0%$ to $86.8%$ . On the ImageCLEF benchmark, the “MedVInT-TE” version of our model achieved a significant improvement with an accuracy rate of $70.5%$ , significantly higher than the previous state-of-the-art (SOTA) accuracy of $62.4%$ .
需要注意的是,VQA-RAD和SLAKE数据集中的问题均分为开放式和封闭式两类。封闭式问题将答案限定在预定义的选项集中,而开放式问题则允许自由文本回答。具体而言,在开放式问题上,准确率从VQA-RAD的$67.2%$提升至$73.7%$,SLAKE数据集上从$81.9%$提升至$88.2%$。对于封闭式问题,MedVInT模型将准确率从$84.0%$提高至$86.8%$。在ImageCLEF基准测试中,模型的"MedVInT-TE"版本取得了显著进步,准确率达到$70.5%$,明显优于此前最先进(SOTA)的$62.4%$准确率。
Beyond comparing baselines with their default settings, we also consider an architecture-specific comparison where all models are directly trained from scratch on the downstream tasks. To distinguish from the default setting, our models here are denoted as “MedVInT-TE-S” and “MedVInT-TD-S”. As shown by the results, our proposed two variants can both surpass the former “M3AE” and “PMC-CLIP” architectures in most cases.
除了比较基线模型及其默认设置外,我们还考虑了针对特定架构的比较,即所有模型均在下游任务上从头开始直接训练。为区别于默认设置,此处我们的模型标记为"MedVInT-TE-S"和"MedVInT-TD-S"。结果表明,在大多数情况下,我们提出的两种变体都能超越之前的"M3AE"和"PMC-CLIP"架构。
Additionally, when comparing the performance of the MedVInT model with and without pre-training on the PMC-VQA-train dataset, using the same architectural framework, it becomes evident that pre-training plays a crucial role in enhancing model performance. Specifically, the “MedVInT-TE” version, when pre-trained, showed a remarkable increase of approximately 16% in accuracy for open-ended questions on VQA-RAD and a 4% increase on SLAKE, compared to the “MedVInT-TE-S” version, which denotes training the model from scratch. Similar enhancements were observed with the “MedVInT-TD” version.
此外,在比较MedVInT模型在PMC-VQA-train数据集上有无预训练的性能时(使用相同的架构框架),可以明显看出预训练对提升模型性能起着关键作用。具体而言,经过预训练的"MedVInT-TE"版本在VQA-RAD开放性问题上的准确率比从零开始训练的"MedVInT-TE-S"版本提高了约16%,在SLAKE上提高了4%。"MedVInT-TD"版本也观察到了类似的提升。
2.3 Evaluation on PMC-VQA
2.3 PMC-VQA评估
In this section, we introduce a new MedVQA benchmark, termed as PMC-VQA-test. We evaluate different models for both open-ended (Blanking) and multiple-choice (Choice) tasks. The results are summarized in Table 3. GPT-4-Oracle refers to the use of GPT-4 to answer questions based on the original captions of figures in academic papers. This approach represents the upper bound of model performance, as it leverages the most accurate and comprehensive information available about each figure. As shown in the tables, when only using language, the model is unable to provide accurate answers and give nearly random outcomes, with an accuracy of only $27.2%$ in Blanking and $30.8%$ in Choice for LLaMA and enhancing the language model from LLaMA to latest GPT-4 still cannot improve the results, i.e., $21.1%$ in Blanking and $25.7%$ in Choice for GPT-4. The lower score in Blanking is due to the language model’s tendency to output longer sentences that cannot be correctly matched to a specific choice, which affects the calculation of model’s accuracy. It is worth noting that around $30%$ of the questions have “B” answers, making the $30.8%$ score nearly equivalent to the highest possible score attainable through guessing. These observations highlight the crucial requirement of multimodal understanding in our dataset and emphasize the strong relationship between images and the questions posed. In contrast to the training split, PMC-VQA-test has undergone thorough manual checking (Check Sec. 4.1 for more details), ensuring the credibility of the evaluation. We also report the experimental results on the original randomly split test set PMC-VQA-test-initial, which is larger but lacks further manual checking, in the supplementary materials A.2.
在本节中,我们介绍了一个名为PMC-VQA-test的新MedVQA基准测试。我们评估了不同模型在开放式(Blanking)和多项选择(Choice)任务上的表现,结果总结在表3中。GPT-4-Oracle指利用GPT-4基于学术论文中图片原始标题回答问题的方案,这种方法代表了模型性能的上限,因为它利用了每张图片最准确全面的信息。如表所示,当仅使用语言时,模型无法提供准确答案且输出近乎随机结果——LLaMA在Blanking任务中准确率仅为$27.2%$,Choice任务中为$30.8%$;即使将语言模型从LLaMA升级至最新的GPT-4仍无法提升结果(GPT-4在Blanking中为$21.1%$,Choice中为$25.7%$)。Blanking得分较低是因为语言模型倾向于输出无法与特定选项匹配的长句,从而影响准确率计算。值得注意的是约$30%$的问题答案为"B",这使得$30.8%$的得分几乎等同于通过猜测可能获得的最高分。这些发现凸显了我们数据集中多模态理解的关键需求,并强调了图像与问题之间的强关联性。与训练集不同,PMC-VQA-test经过彻底人工核查(详见第4.1节),确保了评估的可信度。我们还在补充材料A.2中报告了原始随机划分测试集PMC-VQA-test-initial的实验结果,该数据集规模更大但未经过额外人工核查。
Table 2 | Comparison of ACC to SOTA approaches on VQA-RAD, SLAKE, and ImageClef-VQA-2019. We use the blank model for evaluation which provides output as free text answers rather than multiple-choice options. Pre-training data indicates whether the model is pre-trained on the medical multi-modal dataset before training on the target dataset. “MedVInT-TE-S” and “MedVInT-TD-S” respectively denotes we train the same architecture as “MedVInT-TE” or “MedVInT-TD” from scratch without pre-training on PMC-VQA. The best result is bold, the second-best result is underlined.
| Method | PretrainingData | VQA-RAD | SLAKE | VQA-2019 | ||
| Open | Close | Open | Close | Overall | ||
| M3AE | 66.5 | 79.0 | 79.2 | 83.4 | ||
| PMC-CLIP | 52.0 | 75.4 | 72.7 | 80.0 | ||
| MedVInT-TE-S | 53.6 (41.3,64.8) | 76.5 (69.1,84.9) | 84.0 (80.4,88.4) | 85.1 (79.3,90.1) | 67.9 (60.6,74.2) | |
| MedVInT-TD-S | 55.3 (45.4,69.4) | 80.5 (74.3,89.4) | 79.7 (74.6,85.3) | 85.1 (78.2,89.3) | 58.4 (50.6,66.2) | |
| Hanlin | Unknown | 62.4 | ||||
| MEVF-BAN | VQA-RAD* [28] | 49.2 | 77.2 | 77.8 | 79.8 | |
| CPRD-BAN | ROCO,MedICaT[46,52] | 52.5 | 77.9 | 79.5 | 83.4 | |
| M3AE | CC12M [9] | 67.2 | 83.5 | 80.3 | 87.8 | |
| PMC-CLIP | PMC-OA [32] | 67.0 | 84.0 | 81.9 | 88.0 | |
| MedVInT-TE | PMC-VQA | 69.3 (55.9,79.3) | 84.2 (76.8,90.4) | 88.2 (84.6,92.7) | 87.7 (81.3,92.8)70.5 (62.8,78.2) | |
| MedVInT-TD | PMC-VQA | 73.7(64.8,84.5) | 86.8(80.4,95.5) | 84.5 (80.4,90.5) | 86.3 (79.6,90.6) | 61.0 (53.0,67.6) |
* “Hanlin” is a solution in VQA-2019 challenge instead of a detailed scientific paper and, thus, no more details are provided. The numbers are directly copied from challenge papers. “MEVF-BAN” views the images in the train set of VQA-RAD as a pre training dataset, performs image-wise self-supervised learning on it, and finetunes the model with VQA cases on each dataset. . We utilize the results of MEVF-BAN on various VQA benchmarks as reported by PMC-CLIP.
表 2 | 在VQA-RAD、SLAKE和ImageClef-VQA-2019上ACC与SOTA方法的对比。我们使用空白模型进行评估,该模型提供自由文本答案而非多项选择输出。预训练数据表明模型是否在目标数据集训练前经过医学多模态数据集预训练。"MedVInT-TE-S"和"MedVInT-TD-S"分别表示我们从头开始训练与"MedVInT-TE"或"MedVInT-TD"相同的架构,而未在PMC-VQA上进行预训练。最佳结果加粗,次佳结果加下划线。
| 方法 | 预训练数据 | VQA-RAD-开放 | VQA-RAD-封闭 | SLAKE-开放 | SLAKE-封闭 | VQA-2019总体 |
|---|---|---|---|---|---|---|
| M3AE | 66.5 | 79.0 | 79.2 | 83.4 | ||
| PMC-CLIP | 52.0 | 75.4 | 72.7 | 80.0 | ||
| MedVInT-TE-S | 53.6 (41.3,64.8) | 76.5 (69.1,84.9) | 84.0 (80.4,88.4) | 85.1 (79.3,90.1) | 67.9 (60.6,74.2) | |
| MedVInT-TD-S | 55.3 (45.4,69.4) | 80.5 (74.3,89.4) | 79.7 (74.6,85.3) | 85.1 (78.2,89.3) | 58.4 (50.6,66.2) | |
| Hanlin | Unknown | 62.4 | ||||
| MEVF-BAN | VQA-RAD* [28] | 49.2 | 77.2 | 77.8 | 79.8 | |
| CPRD-BAN | ROCO,MedICaT [46,52] | 52.5 | 77.9 | 79.5 | 83.4 | |
| M3AE | CC12M [9] | 67.2 | 83.5 | 80.3 | 87.8 | |
| PMC-CLIP | PMC-OA [32] | 67.0 | 84.0 | 81.9 | 88.0 | |
| MedVInT-TE | PMC-VQA | 69.3 (55.9,79.3) | 84.2 (76.8,90.4) | 88.2 (84.6,92.7) | 87.7 (81.3,92.8) | 70.5 (62.8,78.2) |
| MedVInT-TD | PMC-VQA | 73.7 (64.8,84.5) | 86.8 (80.4,95.5) | 84.5 (80.4,90.5) | 86.3 (79.6,90.6) | 61.0 (53.0,67.6) |
- "Hanlin"是VQA-2019挑战赛中的一个解决方案而非详细科学论文,因此未提供更多细节。数据直接来自挑战赛论文。"MEVF-BAN"将VQA-RAD训练集中的图像视为预训练数据集,对其进行图像级自监督学习,并在每个数据集上用VQA案例微调模型。我们采用PMC-CLIP报告的各种VQA基准测试中MEVF-BAN的结果。
We also present the zero-shot evaluation results of the general VQA models like PMC-CLIP, BLIP-2, and Open-Flamingo which show relatively lower performance on the choice task. For instance, in the choice task, the model Open-Flamingo only achieved a $26.4%$ accuracy rate, significantly lower performance than our model at $40.3%$ . We also evaluate the medical-specific generative-based VQA model, e.g., LLaVA-Med. Though it is better than the general models, it still lags behind our proposed MedVinT. It’s worth noting that LLaVA-Med is a work after our first announcement. This contrasts with the trained models on PMC-VQA, where we see notable improvements. Specifically, the MedVInT-TE and MedVInT-TD models, when paired with the PMC-CLIP vision backbone, demonstrate superior performance. For the open-ended task, the PMC-CLIP vision backbone again proves beneficial, with the MedVInT-TE model reaching the highest accuracy $(36.4%)$ and BLEU-1 score $(23.2%)$ when combined with the PubMedBERT language backbone. Moreover, the comparison between models trained from scratch and those utilizing CLIP or PMC-CLIP as vision backbones across different configurations of language backbones (PubMedBERT, LLaMA-ENC, and PMC-LLaMA-ENC) reveals a consistent trend: pre-trained models, especially those pre-trained with domainspecific data (PMC-CLIP), tend to outperform their counterparts trained from scratch. This emphasizes the importance of pre-training in achieving higher accuracies and better natural language generation metrics in MedVQA tasks. We then prompted a Large Language Model (LLM) to answer questions based on these generated captions. We also compared our approach with two-stage Visual Question Answering (VQA) models, which employ image captioning followed by a large language model for question answering. We experimented with a two-stage VQA method similar to Chatcad [55]. We first used MedICap [41], a state-of-the-art medical image captioning model, to interpret the given images into captions. The results showed poor performance on the test set. We then trained MedICap on the original image-caption pairs from the PMC-VQA training set to mitigate the domain gap. As shown, MedICap-PMCVQA-GPT-4 still shows inferior performance, which highlights key challenges in the two-stage approach: Captioning models need to anticipate potential questions in their descriptions. There’s often a mismatch between caption content and question focus. For example, a caption might state “This is an MRI image of a brain.” while the question asks “Is there a mass in the image?”. To provide a more comprehensive understanding of the dataset, we offer additional examples illustrated in
我们还展示了通用视觉问答(VQA)模型如PMC-CLIP、BLIP-2和Open-Flamingo的零样本评估结果,这些模型在选择题任务上表现相对较低。例如,在选择题任务中,Open-Flamingo模型仅达到26.4%的准确率,显著低于我们模型的40.3%。我们还评估了基于生成的医疗专用VQA模型LLaVA-Med。虽然它优于通用模型,但仍落后于我们提出的MedVinT。值得注意的是,LLaVA-Med是在我们首次公布之后的工作。这与在PMC-VQA上训练的模型形成对比,我们看到了显著的改进。具体而言,MedVInT-TE和MedVInT-TD模型与PMC-CLIP视觉骨干配对时表现出卓越性能。对于开放式任务,PMC-CLIP视觉骨干再次证明有益,MedVInT-TE模型与PubMedBERT语言骨干结合时达到最高准确率(36.4%)和BLEU-1分数(23.2%)。此外,从头训练的模型与使用CLIP或PMC-CLIP作为视觉骨干的模型在不同语言骨干(PubmedBERT、LLaMA-ENC和PMC-LLaMA-ENC)配置下的比较揭示了一致趋势:预训练模型,特别是使用领域特定数据(PMC-CLIP)预训练的模型,往往优于从头训练的对应模型。这强调了预训练在MedVQA任务中实现更高准确率和更好自然语言生成指标的重要性。我们随后提示一个大语言模型(LLM)基于这些生成的描述回答问题。我们还将我们的方法与两阶段视觉问答(VQA)模型进行了比较,这些模型采用图像描述生成后接大语言模型进行问答。我们实验了类似于Chatcad[55]的两阶段VQA方法。我们首先使用最先进的医学图像描述模型MedICap[41]将给定图像解释为描述。结果显示在测试集上表现不佳。然后我们在PMC-VQA训练集的原始图像-描述对上训练MedICap以减少领域差距。如图所示,MedICap-PMCVQA-GPT-4仍然表现较差,这凸显了两阶段方法的关键挑战:描述模型需要在其描述中预判潜在问题。描述内容与问题焦点之间经常存在不匹配。例如,描述可能说"这是一张脑部MRI图像",而问题问"图像中有肿块吗?"。为了更全面地理解数据集,我们提供了额外的示例说明
Table 3 | Comparison of baseline models using different pre-trained models on both open-ended (Blank) and multiple-choice (Choice) tasks. We reported the results of the PMC-VQA-test. “Scratch” means to train the vision model from scratch with the same architecture as PMC-CLIP.
| Method | Language Backbone | VisionBackbone | Choice | Blanking | ||
| ACC | ACC | BLEU-1 | ||||
| Language-only | ||||||
| GPT-4-Oracle [43] | GPT-4 [43] | 89.3 (87.7,90.8) | 22.0 (19.6,24.5) | 18.8 (17.6,20.2) | ||
| GPT-4 [43] | GPT-4 [43] | 25.7 (23.5,28.1) | 21.1(18.8,23.5) | 3.0(2.6,3.4) | ||
| LLaMA [54] | LLaMA [54] | 30.8 (27.4,34.8) | 27.2 (23.1,31.3) | 14.6 (12.7,16.6) | ||
| Zero-shot | ||||||
| PMC-CLIP [32] | PMC-CLIP [32] | PMC-CLIP [32] | 24.7 (21.3,28.0) | |||
| BLIP-2 [30] | OPT-2.7B [63] | CLIP [47] | 24.3 (20.7,27.7) | 21.8 (17.2,26.4) | 7.6 (5.3,9.9) | |
| Open-Flamingo [4] | LLaMA [54] | CLIP [47] | 26.4 (22.7,29.8) | 26.5 (22.3,30.7) | 4.1 (2.1,6.13) | |
| LLaVA-Med [29] | Vicuna [14] | BioMedCLIP [64] | 34.8 (32.2,37.8) | 29.4 (26.6,32.1) | 3.9(3.5,4.2) | |
| MedICap-GPT-4 | GPT-4 [43] | MedICap [41] | 27.2 (24.7,29.7) | 20.9 (18.8,23.3) | 4.2 (3.6,4.6) | |
| Trained on PMC-VQA | ||||||
| MedICap-PMCVQA-GPT-4 GPT-4 [43] | MedICap-PMCVQA | 35.9 (33.0,38.3) | 22.4 (20.1,24.8) | 3.8 (3.3,4.3) | ||
| MedVInT-TE | Scratch | 34.9 (31.7,38.5) | 34.2 (31.2,37.0) | 20.9 (18.9,23,2) | ||
| PubMedBERT [20] | CLIP [47] | 34.3 (30.7,37.8) | 34.4 (31.0,37.6) | 20.8 (18.6,23.3) | ||
| PMC-CLIP [32] | 37.6 (34.7,40.9) | 36.4 (32.6,39.4) | 23.2 (21.2,25.7) | |||
| LLaMA-ENC [54] | Scratch | 35.2 (31.8,38.3) | 32.5 (29.6,35.9) | 15.9 (12.8,16.8) | ||
| CLIP [47] | 36.1 (31.0,39.5) | 33.4 (29.8,36.5) | 15.1 (12.8,17.5) | |||
| PMC-CLIP [32] | 37.1 (34.0,40.1) | 36.8 (33.5,40.0) | 18.4 (15.6,20.5) | |||
| PMC-LLaMA-ENC [57] | Scratch | 38.0 (34.9,42.2) | 35.0 (31.9,38.5) | 17.0 (14.5,18.9) | ||
| CLIP [47] | 38.5 (35.7,42.4) | 34.4 (31.3,37.8) | 16.5 (14.4,18.8) | |||
| PMC-CLIP [32] | 39.2 (36.7,41.7) | 35.3 (31.4, 38.8) | 18.6 (16.6,21.6) | |||
| MedVInT-TD | LLaMA [54] | Scratch | 37.9 (34.5,41.4) | 30.2 (26.9,33.8) | 18.0 (16.2,20.0) | |
| CLIP [47] | 39.2 (35.3,42.7) | 32.2 (29.4,36.0) | 20.0 (17.8,23.0) | |||
| PMC-CLIP [32] | 39.5 (35.1,42.7) | 33.4 (30.6,37.4) | 21.3 (18.9,23.8) | |||
| Scratch | 36.9 (33.2,40.2) | 29.8 (26.9,32.7) | 17.4 (15.1,19.6) | |||
| PMC-LLaMA [57] | CLIP [47] | 36.9 (32.9,40.1) | 32.6 (29.0,36.2) | 20.4 (18.1,22.9) | ||
| PMC-CLIP [32] | 40.3 (37.2,43.8) | 33.6 (29.9,36.5) | 21.5 (19.4,24.0) | |||
表 3 | 不同预训练模型在开放式 (Blank) 和选择题 (Choice) 任务上的基线模型对比。我们报告了 PMC-VQA-test 的结果。"Scratch" 表示使用与 PMC-CLIP 相同架构从头开始训练视觉模型。
| 方法 | 语言主干 | 视觉主干 | Choice ACC | Blanking ACC | Blanking BLEU-1 |
|---|---|---|---|---|---|
| * * 纯语言模型* * | |||||
| GPT-4-Oracle [43] | GPT-4 [43] | 89.3 (87.7,90.8) | 22.0 (19.6,24.5) | 18.8 (17.6,20.2) | |
| GPT-4 [43] | GPT-4 [43] | 25.7 (23.5,28.1) | 21.1 (18.8,23.5) | 3.0 (2.6,3.4) | |
| LLaMA [54] | LLaMA [54] | 30.8 (27.4,34.8) | 27.2 (23.1,31.3) | 14.6 (12.7,16.6) | |
| * * 零样本* * | |||||
| PMC-CLIP [32] | PMC-CLIP [32] | PMC-CLIP [32] | 24.7 (21.3,28.0) | ||
| BLIP-2 [30] | OPT-2.7B [63] | CLIP [47] | 24.3 (20.7,27.7) | 21.8 (17.2,26.4) | 7.6 (5.3,9.9) |
| Open-Flamingo [4] | LLaMA [54] | CLIP [47] | 26.4 (22.7,29.8) | 26.5 (22.3,30.7) | 4.1 (2.1,6.13) |
| LLaVA-Med [29] | Vicuna [14] | BioMedCLIP [64] | 34.8 (32.2,37.8) | 29.4 (26.6,32.1) | 3.9 (3.5,4.2) |
| MedICap-GPT-4 | GPT-4 [43] | MedICap [41] | 27.2 (24.7,29.7) | 20.9 (18.8,23.3) | 4.2 (3.6,4.6) |
| * * 在 PMC-VQA 上训练* * | |||||
| MedICap-PMCVQA-GPT-4 | GPT-4 [43] | MedICap-PMCVQA | 35.9 (33.0,38.3) | 22.4 (20.1,24.8) | 3.8 (3.3,4.3) |
| * * MedVInT-TE* * | |||||
| PubMedBERT [20] | Scratch | 34.9 (31.7,38.5) | 34.2 (31.2,37.0) | 20.9 (18.9,23,2) | |
| CLIP [47] | 34.3 (30.7,37.8) | 34.4 (31.0,37.6) | 20.8 (18.6,23.3) | ||
| PMC-CLIP [32] | 37.6 (34.7,40.9) | 36.4 (32.6,39.4) | 23.2 (21.2,25.7) | ||
| LLaMA-ENC [54] | Scratch | 35.2 (31.8,38.3) | 32.5 (29.6,35.9) | 15.9 (12.8,16.8) | |
| CLIP [47] | 36.1 (31.0,39.5) | 33.4 (29.8,36.5) | 15.1 (12.8,17.5) | ||
| PMC-CLIP [32] | 37.1 (34.0,40.1) | 36.8 (33.5,40.0) | 18.4 (15.6,20.5) | ||
| PMC-LLaMA-ENC [57] | Scratch | 38.0 (34.9,42.2) | 35.0 (31.9,38.5) | 17.0 (14.5,18.9) | |
| CLIP [47] | 38.5 (35.7,42.4) | 34.4 (31.3,37.8) | 16.5 (14.4,18.8) | ||
| PMC-CLIP [32] | 39.2 (36.7,41.7) | 35.3 (31.4,38.8) | 18.6 (16.6,21.6) | ||
| * * MedVInT-TD* * | |||||
| LLaMA [54] | Scratch | 37.9 (34.5,41.4) | 30.2 (26.9,33.8) | 18.0 (16.2,20.0) | |
| CLIP [47] | 39.2 (35.3,42.7) | 32.2 (29.4,36.0) | 20.0 (17.8,23.0) | ||
| PMC-CLIP [32] | 39.5 (35.1,42.7) | 33.4 (30.6,37.4) | 21.3 (18.9,23.8) | ||
| PMC-LLaMA [57] | Scratch | 36.9 (33.2,40.2) | 29.8 (26.9,32.7) | 17.4 (15.1,19.6) | |
| CLIP [47] | 36.9 (32.9,40.1) | 32.6 (29.0,36.2) | 20.4 (18.1,22.9) | ||
| PMC-CLIP [32] | 40.3 (37.2,43.8) | 33.6 (29.9,36.5) | 21.5 (19.4,24.0) |
Figure 3. This figure showcases random instances of the original image and corresponding captions, along with multiple-choice questions generated from them.
图 3: 该图展示了原始图像及对应描述的随机实例,以及基于它们生成的多选题。
2.4 Evaluation of Visual Backbone Performance
2.4 视觉骨干网络性能评估
We conducted additional experiments on standard medical image classification tasks to demonstrate the visual backbone’s performance and its improvement through the VQA pre-training. We evaluated our model on the MedMNIST dataset [61], which provides a diverse set of medical imaging modalities and classification tasks.
我们在标准医学图像分类任务上进行了额外实验,以验证视觉主干网络的性能及其通过VQA预训练获得的提升。我们在MedMNIST数据集[61]上评估了模型性能,该数据集提供了多样化的医学成像模态和分类任务。
As shown in Table 4, our MedVInT models demonstrate competitive performance across all three tasks. Notably, MedVInT-TE achieves the best performance on DermaMNIST and the second-best performance on Pneumonia M NIST and Breast M NIST, only slightly behind PMC-CLIP. The results are impressive considering that MedVInT was pre-trained on only 177K images, compared to PMC-CLIP’s 1.6M image-caption pairs. Our results demonstrate the effectiveness of VQA-based pre-training compared to CLIP-style training. While both approaches aim to align visual and textual information, VQA requires a deeper understanding of the image content to answer specific questions. This difference in training objectives appears to lead to more robust visual representations, as evidenced by our model’s competitive performance despite being trained on significantly fewer images. These results demonstrate that our MedVQA task not only “standardizes” data into QA pairs but also substantially improves the visual backbone’s performance on various medical image classification tasks.
如表4所示,我们的MedVInT模型在所有三项任务中都展现出具有竞争力的性能。值得注意的是,MedVInT-TE在DermaMNIST上取得最佳表现,在PneumoniaMNIST和BreastMNIST上位列第二,仅略逊于PMC-CLIP。考虑到MedVInT仅用17.7万张图像进行预训练,而PMC-CLIP使用了160万张图像-标题对,这一结果令人印象深刻。我们的结果表明,基于视觉问答(VQA)的预训练相比CLIP风格训练更具成效。虽然两种方法都旨在对齐视觉与文本信息,但VQA需要更深入理解图像内容以回答特定问题。这种训练目标的差异似乎能产生更鲁棒的视觉表征——我们的模型在训练数据量显著更少的情况下仍保持竞争力便印证了这一点。这些结果证明,我们的MedVQA任务不仅将数据"标准化"为问答对,还显著提升了视觉主干网络在各类医学图像分类任务中的性能。
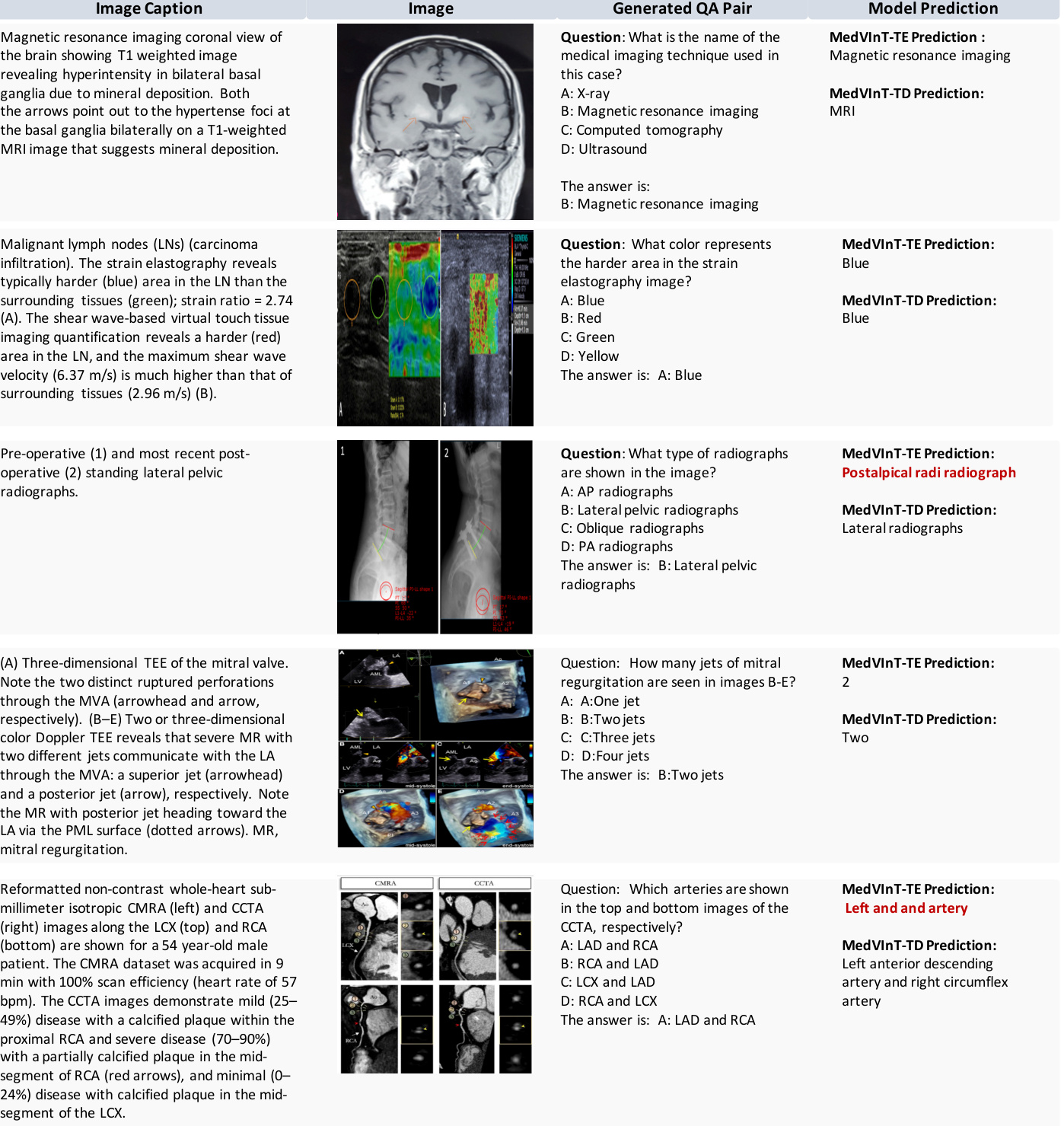
Figure 3 | Examples of image captions, images, the generated question-answer pairs, and model prediction. The wrong predictions are highlighted in red.
图 3 | 图像描述、图像内容、生成的问题-答案对及模型预测结果的示例。错误预测用红色高亮显示。
3 Discussion
3 讨论
In this study, we target the challenge of MedVQA, where even the strongest VQA models trained on natural images yield results that closely resemble random guesses. To overcome this, we propose MedVInT, a generative model tailored to advance this crucial medical task. MedVInT is trained by aligning visual data from a pre-trained vision encoder with language models. Additionally, we present a scalable pipeline for constructing PMC-VQA, a comprehensive VQA dataset in the medical domain comprising 227k pairs across 149k images, spanning diverse modalities and diseases. Our proposed model delivers state-of-the-art performance on existing datasets, providing a new and reliable benchmark for evaluating different methods in this field.
在本研究中,我们针对医学视觉问答 (MedVQA) 的挑战展开研究。即使是在自然图像上训练的最强 VQA 模型,其表现也近乎随机猜测。为此,我们提出了 MedVInT——一种专为推进这一关键医疗任务而设计的生成式模型。MedVInT 通过将预训练视觉编码器的视觉数据与大语言模型对齐进行训练。此外,我们提出了一种可扩展的流程来构建 PMC-VQA,这是医学领域一个全面的 VQA 数据集,包含 149k 张图像中的 227k 个问答对,涵盖多种模态和疾病。我们提出的模型在现有数据集上实现了最先进的性能,为该领域不同方法的评估提供了全新且可靠的基准。
Table 4 | Classification results on three representative subsets of MedMNIST: Pneumonia M NIST (chest X-ray), Breast M NIST (ultrasound), and DermaMNIST (der matos copy). The best results are in bold, and the second-best are in underlined.
| Methods | PneumoniaMNIST | BreastMNIST | DermaMNIST | |||
| AUC↑ | ACC↑ | AUC↑ | ACC↑ | AUC↑ | ACC↑ | |
| ResNet50 [21] | 96.20 | 88.40 | 86.60 | 84.20 | 91.20 | 73.10 |
| DWT-CV [13] | 95.69 | 88.67 | 89.77 | 85.68 | 91.67 | 74.75 |
| SADAE [19] | 98.30 | 91.80 | 91.50 | 87.80 | 92.70 | 75.90 |
| PMC-CLIP | 99.02 | 95.35 | 94.56 | 91.35 | 93.41 | 79.80 |
| MedVInT-TE | 98.49 | 94.87 | 93.44 | 90.38 | 93.71 | 80.00 |
| MedVInT-TD | 97.39 | 94.71 | 90.04 | 87.82 | 93.43 | 78.30 |
表 4 | MedMNIST三个代表性子集的分类结果:PneumoniaMNIST(胸部X光)、BreastMNIST(超声)和DermaMNIST(皮肤镜)。最佳结果以粗体显示,次佳结果以下划线显示。
| 方法 | PneumoniaMNIST | BreastMNIST | DermaMNIST | |||
|---|---|---|---|---|---|---|
| AUC↑ | ACC↑ | AUC↑ | ACC↑ | AUC↑ | ACC↑ | |
| ResNet50 [21] | 96.20 | 88.40 | 86.60 | 84.20 | 91.20 | 73.10 |
| DWT-CV [13] | 95.69 | 88.67 | 89.77 | 85.68 | 91.67 | 74.75 |
| SADAE [19] | 98.30 | 91.80 | 91.50 | 87.80 | 92.70 | 75.90 |
| PMC-CLIP | * * 99.02* * | * * 95.35* * | * * 94.56* * | * * 91.35* * | 93.41 | 79.80 |
| MedVInT-TE | 98.49 | 94.87 | 93.44 | 90.38 | 93.71 | _ 80.00_ |
| MedVInT-TD | 97.39 | 94.71 | 90.04 | 87.82 | 93.43 | 78.30 |
Significance of Medical VQA for Medical Imaging Ecosystem. The development of advanced MedVQA systems has far-reaching implications for various stakeholders in the medical imaging ecosystem [5, 60, 15]. For radiologists and referring physicians, MedVQA can serve as a powerful decision-support tool, potentially enhancing diagnostic precision and streamlining image interpretation processes [16]. This could lead to more efficient clinical workflows and allow healthcare professionals to dedicate more time to direct patient care. For patients, MedVQA systems can significantly improve the communication of complex medical information. By translating intricate radiology reports into more comprehensible language, these systems can enhance patient understanding and engagement in their healthcare journey. This aligns with the growing emphasis on patient-centered care and shared decision-making in modern healthcare practices [45]. From a research and education perspective, MedVQA systems like MedVInT, trained on comprehensive datasets such as PMC-VQA, can serve as valuable tools for medical students and researchers [49]. They can provide interactive learning experiences, assist in the design of research plans, and offer insights into complex medical imaging concepts, thereby contributing to the advancement of medical knowledge and skills.
医学VQA对医学影像生态系统的重要意义。先进的MedVQA系统发展对医学影像生态系统中各利益相关方具有深远影响[5,60,15]。对放射科医师和转诊医师而言,MedVQA可作为强大的决策支持工具,有望提升诊断精确度并优化影像解读流程[16],从而建立更高效的临床工作流,让医疗专业人员能投入更多时间于直接患者护理。对患者而言,MedVQA系统能显著改善复杂医疗信息的传达效果,通过将晦涩的放射学报告转化为更易懂的语言,提升患者在医疗过程中的理解与参与度[45],这符合现代医疗实践中日益重视的"以患者为中心"护理和共同决策理念。在科研教育层面,基于PMC-VQA等综合数据集训练的MedVInT等系统,可为医学生和研究人员提供宝贵工具[49],既能创造交互式学习体验,辅助研究方案设计,又能帮助理解复杂医学影像概念,从而推动医学知识与技能的进步。
PMC-VQA Act as a Valuable Resource for Medical VQA Domain. Previous MedVQA datasets are usually limited in size and diversity, as demonstrated in Table 1. In contrast, PMC-VQA represents a pivotal advancement, offering an extensive resource that addresses the diverse and complex needs of the medical VQA domain. PMC-VQA facilitates the development of models capable of understanding and interpreting medical imagery with unprecedented accuracy and detail. Moreover, comparing results using the same architecture, with and without PMC-VQA (Table 3), it is clear that pre-training with PMC-VQA significantly outperforms. These results highlight the critical role that our PMC-VQA plays in addressing the major challenges that hinder the development of a generative MedVQA system. The pre-training enables models to gain a deep understanding of medical visuals and their associated questions, significantly enhancing their predictive capabilities.
PMC-VQA成为医疗VQA领域的宝贵资源。现有MedVQA数据集通常在规模和多样性上存在局限,如表1所示。相比之下,PMC-VQA标志着关键性突破,提供了满足医疗VQA领域复杂多样化需求的丰富资源。PMC-VQA能推动开发具备前所未有的精确医疗图像理解能力的模型。通过对比相同架构下使用与不使用PMC-VQA的实验结果(表3),可明确观察到PMC-VQA预训练带来的显著性能提升。这些结果凸显了PMC-VQA在解决阻碍生成式MedVQA系统发展的核心挑战中的关键作用。预训练使模型能深入理解医学视觉内容及其相关问题,从而大幅提升预测能力。
General Visual-language Models Struggle on MedVQA. We evaluated the zero-shot performance of existing SOTA multimodal models, BLIP-2 and open-source version of Flamingo [30, 4]. As shown, even the best-performing models in natural images struggle to answer our questions, demonstrating the challenging nature of our dataset and its strong biomedical relevance. These results highlight the critical role that our PMC-VQA-train plays in addressing the major challenges that hinder the development of a generative MedVQA system.
通用视觉语言模型在MedVQA任务中表现欠佳。我们评估了现有SOTA多模态模型BLIP-2和开源版Flamingo [30,4]的零样本性能。结果显示,即便在自然图像领域表现最优的模型也难以正确回答我们的问题,这证明我们的数据集具有显著挑战性及高度生物医学相关性。这些结果凸显了PMC-VQA-train数据集在解决阻碍生成式MedVQA系统发展的关键挑战中所起的重要作用。
MedVInT Achieves State-of-the-art Performance of Generative MedVQA. As demonstrated in the results, both MedVInT-TE and MedVInT-TD perform well on the MedVQA tasks. We compared it against various baselines that use different generative model backbones. Our results show that replacing the general visual backbone with a specialized medical one leads to improved performance, highlighting the importance of visual understanding in our test set. Additionally, we observed that replacing the language backbone with a domain-specific model also leads to some improvements, although not as significant as those achieved in the visual domain. In addition, the gap between the two training styles mainly exists in open-ended questions, with “MedVInT-TD” performing better on VQA-RAD and “MedVInT-TE” being more effective on SLAKE. This difference can be attributed to the fact that the VQA-RAD answers are typically longer than those in SLAKE, making the “MedVInT-TD” model more suitable. Conversely, SLAKE questions often require short responses, making the “MedVInT-TE” model more appropriate for such retrieve-like tasks.
MedVInT实现生成式医学视觉问答(MedVQA)最先进性能。结果显示,MedVInT-TE和MedVInT-TD在MedVQA任务中均表现优异。我们对比了使用不同生成模型基线的多种基准方法,结果表明:用专业医学视觉主干网络替代通用视觉主干可提升性能,这凸显了视觉理解在我们测试集中的重要性。此外,将语言主干替换为领域专用模型也能带来一定改进,但效果不如视觉领域显著。两种训练方式的差异主要体现在开放式问题上:MedVInT-TD在VQA-RAD数据集表现更优,而MedVInT-TE在SLAKE数据集更有效。这种差异源于VQA-RAD的答案通常比SLAKE更长,使得MedVInT-TD模型更适用;相反,SLAKE问题常需简短回答,因此MedVInT-TE模型更适合这类检索型任务。
PMC-VQA-test Presents a Significantly More Challenging Benchmark. Notably, the previous SOTA medical multimodal model, PMC-CLIP [32], struggles on our dataset. Not only does it fail to solve the blanking task, but it also significantly under performs on multi-choice questions, with accuracy close to random. These findings underline the difficulty of our proposed benchmark and its capacity to provide a more rigorous evaluation of VQA models. However, while evaluating our proposed challenging benchmark, even the state-of-the-art models struggle, showing that there is still ample room for development in this field.
PMC-VQA-test提出了一个显著更具挑战性的基准。值得注意的是,此前最先进的医疗多模态模型PMC-CLIP [32]在我们的数据集上表现不佳。它不仅无法解决填空任务,在多项选择题上的准确率也显著低于预期,接近随机猜测水平。这些发现凸显了我们提出的基准任务的难度,以及其对VQA模型进行更严格评估的能力。然而,在评估这一挑战性基准时,即使是当前最先进的模型也表现吃力,表明该领域仍有广阔的发展空间。
Impacts of Our Work. Since released to the public, we are delighted to observe the rapid adoption and extensive utilization of the PMC-VQA dataset, across a diverse range of research endeavors since its release. The dataset has served as a foundational resource for the development of numerous generative models, demonstrating its significant impact on the field. Notable examples include MathVista [38], RadFM [58], Qilin-Med-VL [36], SILKIE [31], CheXagent [12], UniDCP [62], and Quilt-LLaVA [50]. In addition, the methodology employed in constructing the dataset and the innovative prompt strategies we introduced have also inspired a series of works [59] and [10]. Furthermore, many studies have compared with our proposed MedVInT, recognizing it as the pioneering medical generative foundation model, such as Med-flamingo [39], OmniMedVQA [23]. This widespread adoption not only validates the robustness and utility of our dataset but also highlights its role in the scientific community.
我们工作的影响。自公开发布以来,我们欣喜地观察到PMC-VQA数据集在各类研究中的快速采用和广泛应用。该数据集已成为开发众多生成式模型的基础资源,展现了其对领域的重大影响。代表性成果包括MathVista [38]、RadFM [58]、Qilin-Med-VL [36]、SILKIE [31]、CheXagent [12]、UniDCP [62]和Quilt-LLaVA [50]。此外,数据集构建方法和我们提出的创新提示策略也启发了系列研究[59][10]。更多研究将我们提出的MedVInT视为医疗生成式基础模型的开创性工作进行比较,例如Med-flamingo [39]、OmniMedVQA [23]。这种广泛采用不仅验证了我们数据集的稳健性和实用性,更凸显了其在科学界的重要作用。
Limitations. The proposed PMC-VQA, while comprehensive, is subject to several limitations. First, similar to all existing datasets, there might be potential distribution biases in the images included in PMC-VQA compared to clinical practice. Specifically, our data is curated from academic papers, where there may be selective use of images to illustrate typical cases or slices, along with additional annotations such as arrows to aid understanding, resulting in our data being simpler compared to clinical scenarios. Nevertheless, for training purposes, the data from PMC-VQA remains crucial to help models better understand real clinical imaging data, as shown by the performance on public benchmarks in Table 2. On the other hand, for testing, i.e., the benchmark we propose as shown in Table 3, even in such relatively simple scenarios, current methods still face significant challenges. Hence, for the ongoing advancement of MedVQA, conducting assessments in such an experimental playground to steer the emergence of more potent methodologies for the future still holds significance. On evaluation metrics, measuring the results from generative models poses a general challenge in the entire AI community [14], and this holds true for our testing as well. Although both the ACC score and Bleu score are used in our benchmark for assessing open-ended blanking results, these two metrics fail to capture the fluency of the generated sentence since they measure string similarity irrespective of word order. The encoder-based model thus significantly under performs the decoder-based model in this regard. To address this issue, we plan to explore more accurate and effective evaluation metrics in our benchmark in future work. Lastly, as a starting point for generative-based MedVQA methods, our models may still suffer from hallucinations in non-sensical or adversarial cases with huge domain gaps (more case studies in our supplementary). Thus this paper is more as a proof-of-concept for building generative-based medical VQA models and needs more future efforts for real clinical applications.
局限性。尽管提出的PMC-VQA具有全面性,但仍存在若干局限。首先,与所有现有数据集类似,PMC-VQA包含的图像相比临床实践可能存在潜在分布偏差。具体而言,我们的数据源自学术论文,其中可能选择性使用图像来展示典型病例或切片,并辅以箭头等辅助理解的额外标注,导致数据比临床场景更为简化。然而就训练目的而言,如表2所示,PMC-VQA数据对于帮助模型更好理解真实临床影像数据仍至关重要。另一方面,就测试而言(即表3所示我们提出的基准),即便在此类相对简化的场景中,现有方法仍面临重大挑战。因此,为推动MedVQA的持续发展,在此类实验场中进行评估以引导未来更强力方法的出现仍具意义。在评估指标方面,衡量生成模型的结果是整个AI领域面临的普遍挑战[14],我们的测试亦不例外。虽然基准中同时采用ACC分数和Bleu分数评估开放式填空结果,但这两种指标因仅衡量字符串相似度而忽略词序,无法捕捉生成句子的流畅性。基于编码器的模型在此方面显著逊于基于解码器的模型。为解决该问题,我们计划在未来工作中探索更精准有效的评估指标。最后,作为基于生成的MedVQA方法的起点,我们的模型在存在巨大领域差距的无意义或对抗性案例中仍可能出现幻觉(补充材料含更多案例研究)。因此本文更多是构建基于生成的医疗VQA模型的概念验证,实际临床应用仍需未来更多努力。
4 Method
4 方法
4.1 The PMC-VQA Dataset
4.1 PMC-VQA数据集
Our study has identified the lack of large-scale, multi-modal MedVQA datasets as a significant obstacle to the development of effective generative MedVQA models. In this section, we provide a detailed description of our dataset collection process, starting with the source data and continuing with the question-answer generation and data filtering procedures. Finally, we analyze the collected data from various perspectives to gain insights into its properties and potential applications. The main data collection flow can be found in Figure 4.
我们的研究发现,缺乏大规模多模态医学视觉问答(MedVQA)数据集是开发高效生成式MedVQA模型的主要障碍。本节将详细描述数据集收集流程,包括源数据获取、问答生成及数据过滤等步骤,并从多角度分析所收集数据的特性与应用潜力。主要数据收集流程见图4。
Source Data. We start from PMC-OA [32], which is a comprehensive biomedical dataset comprising 1.6 million image-text pairs collected from PubMed Central (PMC)’s OpenAccess subset [48], covering 2.4 million papers. The pipeline of creating PMC-OA consists of three major stages: i medical figure-caption collection; (ii) subfigure separation; (iii) subcaption separation & alignment. To maintain the diversity and complexity of PMC-VQA, we have used a version of 381K image-caption pairs obtained from the first stage of the medical figure collection process without subfigure auto-separation.
源数据。我们从PMC-OA [32]开始,这是一个全面的生物医学数据集,包含从PubMed Central (PMC)开放获取子集[48]收集的160万张图像-文本对,覆盖240万篇论文。创建PMC-OA的流程包含三个主要阶段:(i) 医学图表-标题收集;(ii) 子图分离;(iii) 子标题分离与对齐。为保持PMC-VQA的多样性和复杂性,我们使用了从医学图表收集过程第一阶段获得的38.1万张图像-标题对版本,未进行子图自动分离。
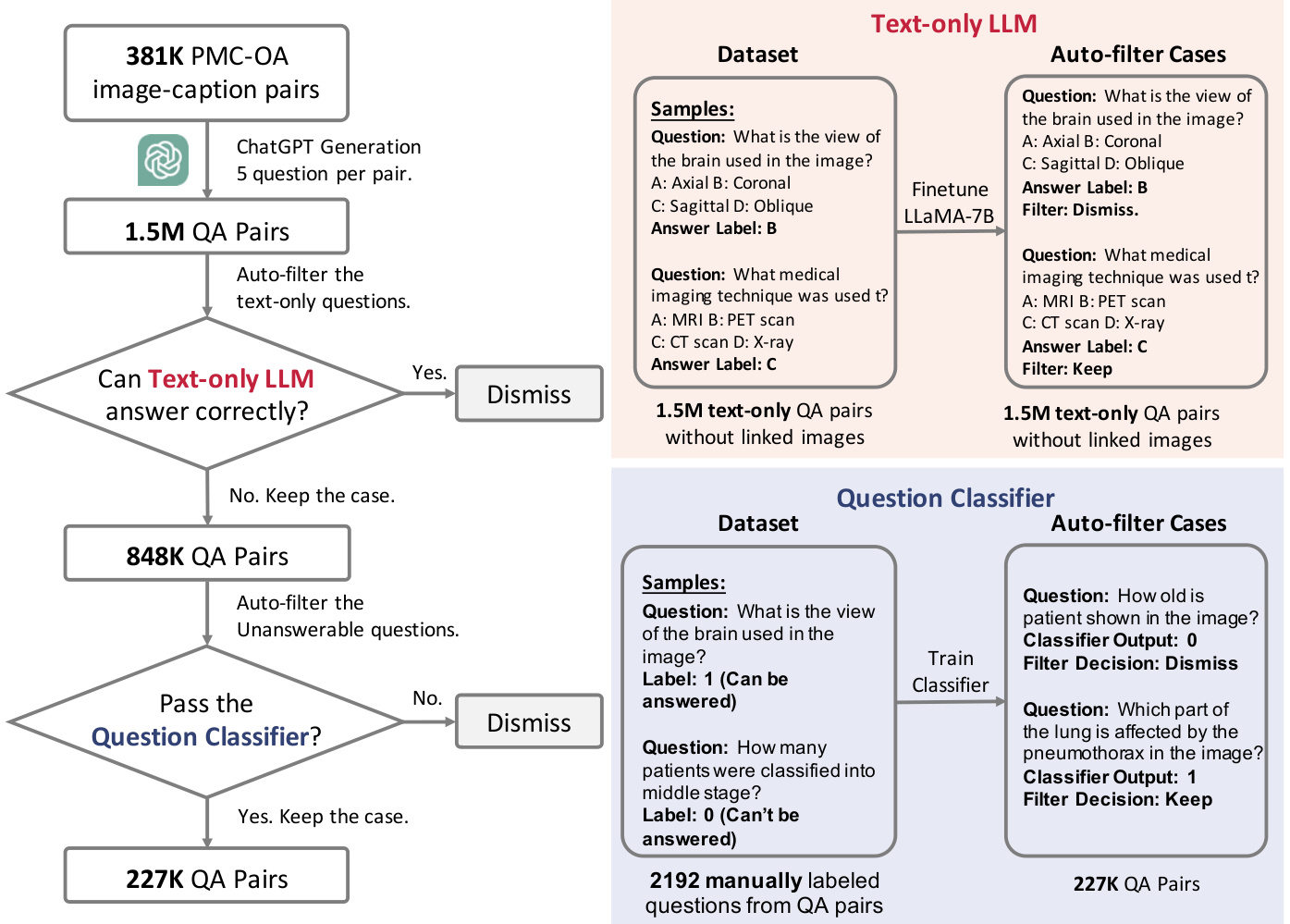
Figure 4 | The whole flowchart demonstrating how we build up our PMC-VQA dataset. In left, we show the general progress and in right we show how we build up the two auto-filter models used in our data collection.
图 4 | 构建PMC-VQA数据集的完整流程图。左侧展示整体流程,右侧展示数据收集中使用的两个自动过滤模型的构建过程。
Question-Answer Generation. To automatically generate high-quality question-answer pairs, we input the image captions of PMC-OA, and prompt ChatGPT to generate 5 question-answer pairs for each caption. We use the following prompt to generate 5 question-answer pairs for each caption.
问答生成。为了自动生成高质量的问答对,我们输入PMC-OA的图像描述,并提示ChatGPT为每个描述生成5个问答对。我们使用以下提示为每个描述生成5个问答对。
Ask 5 questions about the content and generate four options for each question. The questions should be answerable with the information provided in the caption, and the four options should include one correct and three incorrect options, with the position of the correct option randomized. The output should use the following template: i:‘the question index’ question:‘the generate question’ choice: ‘A:option content B:option content C:option content D:option content’ answer: The correct option(A\B\C\D).
i:'1' question:'根据说明,输出模板中哪个部分表示正确答案的位置?' choice: 'A:question B:choice C:answer D:i' answer: The correct option(C).
i:'2' question:'每个问题应生成几个选项?' choice: 'A:2个 B:3个 C:4个 D:5个' answer: The correct option(C).
i:'3' question:'问题内容应基于什么信息来回答?' choice: 'A:标题提供的信息 B:正文提供的信息 C:脚注提供的信息 D:图注提供的信息' answer: The correct option(D).
i:'4' question:'正确选项的位置应该如何设置?' choice: 'A:始终在A位置 B:始终在B位置 C:随机分布 D:始终在D位置' answer: The correct option(C).
i:'5' question:'输出模板中哪个字段用于标识问题序号?' choice: 'A:question B:choice C:answer D:i' answer: The correct option(D).
This approach allows us to generate a large volume of diverse and high-quality questions that cover a wide range of medical topics. Considering some captions are too short to ask 5 questions, ChatGPT will repeat generated question-answer pairs or refuse to generate new pairs halfway and we dismissed the dummy cases. After generating the question-answer pairs using ChatGPT, we applied a rigorous filtering process to ensure that the pairs met our formatting requirements. As a result, we obtained 1,497,808 question-answer pairs, and since the original captions are linked with images, the pairs can naturally find corresponding images, resulting in an average of 3.93 pairs per image.
该方法使我们能够生成大量涵盖广泛医学主题的多样化高质量问题。考虑到部分标题过短无法生成5个问题,ChatGPT会重复生成问答对或在过程中拒绝生成新对,我们剔除了这些无效案例。通过ChatGPT生成问答对后,我们实施了严格的筛选流程以确保其符合格式要求。最终获得1,497,808组问答对,由于原始标题与图像关联,这些问答对可自然匹配对应图像,平均每张图像对应3.93组问答对。
Automatic & Manual Data Filtering. As the questions are sourced from image captions, some of them can be answered correctly using biomedical knowledge alone, i.e., without the need for a specific image, for example, question: “which type of MRI sequence shows high signal in the marrow edema?”. To address this issue, we trained a question-answer model using LLaMA-7B [54] with text data only and eliminated all questions that could be potentially answered by the language model. Specifically, we first split the dataset into two parts, then we train a LLaMA-7B model only text input following the full fine-tuning pipeline introduced in PMC-LLaMA [57] in each part and do inference on the other part. To avoid that sometimes language model may make the correct choice by randomly guessing, for each case, we will shuffle the choice list and do inference five times. The questions the language model can make the right choice three times out of five will be dismissed. This filtering process resulted in 848,433 question-answer pairs that are unanswerable by the language-only model.
自动与手动数据过滤。由于问题来源于图像描述,其中部分仅凭生物医学知识即可正确回答(无需特定图像),例如问题:"哪种MRI序列在骨髓水肿中显示高信号?"。为解决此问题,我们使用LLaMA-7B [54]训练了仅基于文本数据的问答模型,并剔除了所有可能被语言模型回答的问题。具体而言,我们先将数据集分为两部分,随后在各部分中按照PMC-LLaMA [57]提出的全微调流程训练仅接受文本输入的LLaMA-7B模型,并对另一部分数据进行推理。为避免语言模型随机猜测正确选项的情况,我们对每个问题的选项列表进行五次随机排序并推理。若语言模型在五次中有三次选择正确,则该问题将被剔除。此过滤流程最终保留了848,433个纯语言模型无法回答的问答对。
Furthermore, some questions in our data rely on additional information in the caption that cannot be answered with only the corresponding image, such as “How many patients were classified into the middle stage?" To identify these questions, we manually annotated 2192 question-answer pairs with binary labels, using ‘1’ for answerable based on images and ‘0’ otherwise. Then we train and evaluate a question classification model on these labeled data, specifically 1752 pairs for training and 440 for testing, and the model can achieve an accuracy of 81.77% on this binary classification task. We then used this model for data cleaning, resulting in a total of 226,946 question-answer pairs corresponding to 149,075 images, termed as PMC-VQA dataset.
此外,我们数据中的部分问题依赖于标题中的附加信息,仅凭对应图像无法回答,例如"有多少患者被归类为中期?" 为识别这类问题,我们手工标注了2192个带二元标签的问答对,用'1'表示可基于图像回答,'0'表示不可回答。随后在这些标注数据上训练并评估了一个问题分类模型(具体使用1752对训练,440对测试),该模型在此二元分类任务中准确率达到81.77%。我们最终使用该模型进行数据清洗,得到共计226,946个问答对(对应149,075张图像),命名为PMC-VQA数据集。
From this cleaned dataset, we randomly selected 50,000 image-question pairs to create an initial test set, PMC-VQA-test-initial. The same image is guaranteed to not appear in both the training and testing sets. Additionally, we manually checked some test samples again, resulting in a small clean test set of 2,000 samples, which were manually verified for quality, termed as PMC-VQA-test, where we mainly consider the following criteria:
从清洗后的数据集中,我们随机选取了50,000个图像-问题对构建初始测试集PMC-VQA-test-initial,确保同一图像不会同时出现在训练集和测试集中。此外,我们再次人工核验了部分测试样本,最终筛选出2,000个经过质量验证的优质样本组成小型洁净测试集PMC-VQA-test,主要考量以下标准:
During this verification procedure, we have estimated that over 80% cases in PMC-VQA-test can be retained.
在此验证过程中,我们估计PMC-VQA测试集中超过80%的案例可被保留。
4.2 Architecture Design
4.2 架构设计
We start with an introduction to the problem of generative medical visual question answering in Sec. 4.2.1, and detail our proposed architecture for generative MedVQA (Figure 5). We mainly focus on leveraging the pre-trained uni-model model to build up a multi-modal generative VQA achi tec ture. Specifically, we offer two model variants, that are tailored to encoder-based and decoder-based language models, respectively, denoted as MedVInT-TE (Sec. 4.2.2) and MedVInT-TD (Sec. 4.2.3).
我们从第4.2.1节开始介绍生成式医学视觉问答 (generative medical visual question answering) 问题,并详细阐述提出的生成式MedVQA架构 (图5)。主要聚焦于利用预训练单模态模型构建多模态生成式VQA架构。具体而言,我们提供了两种模型变体,分别适配基于编码器和基于解码器的大语言模型,记为MedVInT-TE (第4.2.2节) 和 MedVInT-TD (第4.2.3节)。
4.2.1 Problem Formulation
4.2.1 问题表述
MedVQA is a task of answering natural language questions about medical visual content, typically images or videos obtained from medical devices like X-ray, CT, MRI, or microscopy, etc. Specifically, our goal is to train a model that can output the corresponding answer for a given question, which can be expressed as:
MedVQA是一项关于医学视觉内容(通常来自X光、CT、MRI或显微镜等医疗设备的图像或视频)的自然语言问答任务。具体而言,我们的目标是训练一个模型,能够针对给定问题输出相应答案,可表示为:
$$
\hat{a}_ {i}=\Phi_ {\mathrm{MedVQA}}(\mathcal{Z}_ {i},q_ {i};\Theta)=\Phi_ {\mathrm{dec}}(\Phi_ {\mathrm{vis}}(\mathcal{Z}_ {i};\theta_ {\mathrm{vis}}),\Phi_ {\mathrm{text}}(q_ {i};\theta_ {\mathrm{text}});\theta_ {\mathrm{dec}})
$$
$$
\hat{a}_ {i}=\Phi_ {\mathrm{MedVQA}}(\mathcal{Z}_ {i},q_ {i};\Theta)=\Phi_ {\mathrm{dec}}(\Phi_ {\mathrm{vis}}(\mathcal{Z}_ {i};\theta_ {\mathrm{vis}}),\Phi_ {\mathrm{text}}(q_ {i};\theta_ {\mathrm{text}});\theta_ {\mathrm{dec}})
$$
Here, $\hat{a}_ {i}$ refers to the predicted answer, $\mathcal{Z}_ {i}\in\mathbb{R}^{H\times W\times C}$ refers to the visual image, $H,W,C$ are height, width, channel respectively. The posed question and corresponding ground-truth answer in the form of natural language are denoted as $q_ {i}$ and $a_ {i}$ , respectively. $\Theta={\theta_ {\mathrm{vis}},\theta_ {\mathrm{text}},\theta_ {\mathrm{dec}}}$ denote the trainable parameters.
这里,$\hat{a}_ {i}$ 指预测的答案,$\mathcal{Z}_ {i}\in\mathbb{R}^{H\times W\times C}$ 指视觉图像,$H,W,C$ 分别代表高度、宽度和通道数。以自然语言形式提出的问题及其对应的真实答案分别表示为 $q_ {i}$ 和 $a_ {i}$。$\Theta={\theta_ {\mathrm{vis}},\theta_ {\mathrm{text}},\theta_ {\mathrm{dec}}}$ 表示可训练参数。
Existing approaches have primarily treated medical VQA as a classi
