Towards Generalist Biomedical AI
迈向通用生物医学AI
$^{1}$ Google Research, $^2$ Google DeepMind
$^{1}$ Google Research, $^2$ Google DeepMind
Medicine is inherently multimodal, with rich data modalities spanning text, imaging, genomics, and more. Generalist biomedical artificial intelligence (AI) systems that flexibly encode, integrate, and interpret this data at scale can potentially enable impactful applications ranging from scientific discovery to care delivery. To enable the development of these models, we first curate Multi Med Bench, a new multimodal biomedical benchmark. Multi Med Bench encompasses 14 diverse tasks such as medical question answering, mammography and dermatology image interpretation, radiology report generation and sum mari z ation, and genomic variant calling. We then introduce Med-PaLM Multimodal (Med-PaLM M), our proof of concept for a generalist biomedical AI system. Med-PaLM M is a large multimodal generative model that flexibly encodes and interprets biomedical data including clinical language, imaging, and genomics with the same set of model weights. Med-PaLM M reaches performance competitive with or exceeding the state of the art on all Multi Med Bench tasks, often surpassing specialist models by a wide margin. We also report examples of zero-shot generalization to novel medical concepts and tasks, positive transfer learning across tasks, and emergent zero-shot medical reasoning. To further probe the capabilities and limitations of Med-PaLM M, we conduct a radiologist evaluation of model-generated (and human) chest X-ray reports and observe encouraging performance across model scales. In a side-by-side ranking on 246 retrospective chest X-rays, clinicians express a pairwise preference for Med-PaLM M reports over those produced by radiologists in up to $40.50%$ of cases, suggesting potential clinical utility. While considerable work is needed to validate these models in real-world use cases, our results represent a milestone towards the development of generalist biomedical AI systems.
医学本质上是多模态的,涵盖文本、影像、基因组学等丰富的数据形式。能够灵活编码、整合并大规模解读此类数据的通用生物医学人工智能 (AI) 系统,有望实现从科学发现到医疗护理的广泛应用。为促进此类模型的开发,我们首先构建了 Multi Med Bench——一个新型多模态生物医学基准测试集,涵盖医学问答、乳腺X光与皮肤镜图像解读、放射学报告生成与摘要、基因组变异检测等14项多样化任务。随后我们提出 Med-PaLM Multimodal (Med-PaLM M) 作为通用生物医学AI系统的概念验证。这个大型多模态生成模型采用统一权重,可灵活编码和解读临床语言、影像及基因组数据。Med-PaLM M 在 Multi Med Bench 所有任务中达到或超越当前最优水平,多数情况下大幅领先专科模型。我们还观察到该模型展现的零样本医学概念迁移能力、跨任务正向迁移学习以及涌现的零样本医学推理能力。为进一步探究 Med-PaLM M 的能力边界,我们邀请放射科医生对模型生成(及人工撰写)的胸片报告进行评估,发现不同规模的模型均表现出色。在246份回顾性胸片的双盲评估中,临床医生对 Med-PaLM M 报告的偏好比例最高达40.50%,预示其临床潜力。虽然实际应用仍需大量验证工作,我们的成果标志着通用生物医学AI系统发展的重要里程碑。
1 Introduction
1 引言
Medicine is a multimodal discipline. Clinicians routinely interpret data from a wide range of modalities including clinical notes, laboratory tests, vital signs and observations, medical images, genomics, and more when providing care.
医学是一门多模态学科。临床医生在提供诊疗服务时,通常会综合解读来自临床记录、实验室检测、生命体征观察、医学影像、基因组学等多种模态的数据。
Despite significant progress in biomedical AI, most models today are unimodal single task systems [1–3]. Consider an existing AI system for interpreting mammograms [4]. Although the system obtains state-of-the-art (SOTA) performance on breast cancer screening, it cannot incorporate relevant information such as patient health records (e.g., breast cancer gene screening status), other modalities such as MRI, or published medical literature that might help contextual ize, refine, and improve performance. Further, the system’s output is constrained to a pre-specified set of possible classifications. It cannot verbally explain its prediction or engage in a collaborative dialogue to learn from a physician’s feedback. This bounds performance and utility of these narrow, single-task, unimodal, specialist AI systems in real-world applications.
尽管生物医学AI取得了重大进展,但当今大多数模型仍是单模态单任务系统[1-3]。以现有的乳腺X光片解读AI系统为例[4],虽然该系统在乳腺癌筛查上达到了最先进(SOTA)水平,却无法整合患者健康档案(如乳腺癌基因筛查状态)、MRI等其他模态数据,或是可能帮助优化性能的医学文献等关键信息。此外,系统输出被限制在预设分类范围内,既无法用语言解释预测结果,也不能通过协作对话从医生反馈中学习。这些局限制约了这类单一任务、单模态的专业AI系统在实际应用中的性能和效用。
The emergence of foundation models [5] offers an opportunity to rethink the development of medical AI systems. These models are often trained on large-scale data with self-supervised or unsupervised objectives and can be rapidly and effectively adapted to many downstream tasks and settings using in-context learning or few-shot finetuning [6, 7]. Further, they often have impressive generative capabilities that can enable effective human-AI interaction and collaboration. These advances enable the possibility of building a unified biomedical AI system that can interpret multimodal data with complex structures to tackle many challenging tasks. As the pace of biomedical data generation and innovation increases, so will the potential impact of such models, with a breadth of possible downstream applications spanning fundamental biomedical discovery to care delivery.
基础模型 [5] 的出现为重新思考医疗AI系统的开发提供了契机。这些模型通常通过自监督或无监督目标在大规模数据上进行训练,并能通过上下文学习或少样本微调 [6, 7] 快速有效地适配多种下游任务和场景。此外,它们往往具备出色的生成能力,可实现高效的人机交互与协作。这些进步使得构建统一生物医学AI系统成为可能,该系统能解析具有复杂结构的多模态数据以应对众多挑战性任务。随着生物医学数据生成与创新速度的加快,此类模型的潜在影响将持续扩大,其下游应用范围涵盖从基础生物医学研究到医疗服务的广阔领域。
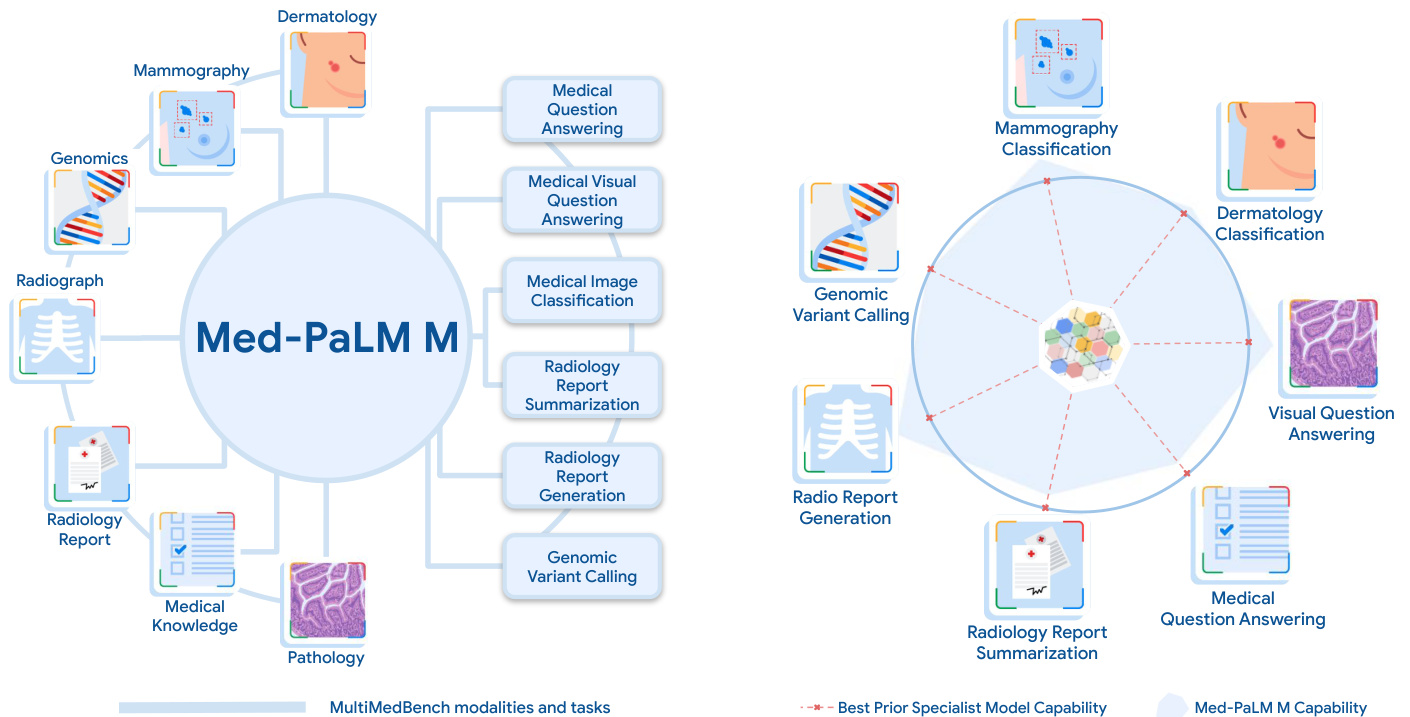
Figure 1 | Med-PaLM M overview. A generalist biomedical AI system should be able to handle a diverse range of biomedical data modalities and tasks. To enable progress towards this over arching goal, we curate Multi Med Bench, a benchmark spanning 14 diverse biomedical tasks including question answering, visual question answering, image classification, radiology report generation and sum mari z ation, and genomic variant calling. Med-PaLM Multimodal (Med-PaLM M), our proof of concept for such a generalist biomedical AI system (denoted by the shaded blue area) is competitive with or exceeds prior SOTA results from specialists models (denoted by dotted red lines) on all tasks in Multi Med Bench. Notably, Med-PaLM M achieves this using a single set of model weights, without any task-specific customization.
图 1 | Med-PaLM M概览。一个通用的生物医学AI系统应当能够处理多种生物医学数据模态和任务。为实现这一总体目标,我们构建了Multi Med Bench基准测试,涵盖14项多样化生物医学任务,包括问答、视觉问答、图像分类、放射学报告生成与摘要以及基因组变异识别。我们的概念验证系统Med-PaLM多模态(Med-PaLM M)(蓝色阴影区域表示)在Multi Med Bench所有任务中均达到或超越专业模型(红色虚线表示)的先前SOTA结果。值得注意的是,Med-PaLM M仅使用单一模型权重就实现了这一目标,无需任何任务特定定制。
In this work, we detail our progress towards such a generalist biomedical AI system - a unified model that can interpret multiple biomedical data modalities and handle many downstream tasks with the same set of model weights. One of the key challenges of this goal has been the absence of comprehensive multimodal medical benchmarks. To address this unmet need, we curate Multi Med Bench, an open source multimodal medical benchmark spanning language, medical imaging, and genomics modalities with 14 diverse biomedical tasks including question answering, visual question answering, medical image classification, radiology report generation and sum mari z ation, and genomic variant calling.
在这项工作中,我们详细介绍了通用生物医学AI系统的研发进展——该统一模型能够解析多种生物医学数据模态,并使用同一组模型权重处理多种下游任务。实现该目标的核心挑战之一是缺乏全面的多模态医学基准测试。为此我们构建了Multi Med Bench:一个开源的多模态医学基准测试集,涵盖语言、医学影像和基因组学模态,包含14项多样化生物医学任务(如问答、视觉问答、医学图像分类、放射学报告生成与摘要、基因组变异检测等)。
We leverage Multi Med Bench to design and develop Med-PaLM Multimodal (Med-PaLM M), a large-scale generalist biomedical AI system building on the recent advances in language [8, 9] and multimodal foundation models [10, 11]. In particular, Med-PaLM M is a flexible multimodal sequence-to-sequence architecture that can easily incorporate and interleave various types of multimodal biomedical information. Further, the expressiveness of the modality-agnostic language decoder enables the handling of various biomedical tasks in a simple generative framework with a unified training strategy.
我们利用Multi Med Bench设计和开发了Med-PaLM Multimodal (Med-PaLM M),这是一个基于语言[8,9]和多模态基础模型[10,11]最新进展的大规模通用生物医学AI系统。具体而言,Med-PaLM M是一种灵活的多模态序列到序列架构,能够轻松整合和交错处理多种类型的多模态生物医学信息。此外,模态无关的语言解码器的表达能力使其能够在简单的生成式框架下,通过统一的训练策略处理各种生物医学任务。
To the best of our knowledge, Med-PaLM M is the first demonstration of a generalist biomedical AI system that can interpret multimodal biomedical data and handle a diverse range of tasks with a single model. Med-PaLM M reaches performance competitive with or exceeding the state-of-the-art (SOTA) on all tasks in Multi Med Bench, often surpassing specialized domain and task-specific models by a large margin. In particular, Med-PaLM M exceeds prior state-of-the-art on chest X-ray (CXR) report generation (MIMIC-CXR dataset) by over 8% on the common success metric (micro-F1) for clinical efficacy. On one of the medical visual question answering tasks (Slake-VQA [12]) in Multi Med Bench, Med-PaLM M outperforms the prior SOTA results by over $10%$ on the BLEU-1 and F1 metrics.
据我们所知,Med-PaLM M 是首个通用生物医学AI系统,能够解读多模态生物医学数据,并用单一模型处理多样化任务。在Multi Med Bench的所有任务中,Med-PaLM M的表现达到或超越当前最优水平(SOTA),且通常大幅领先于专用领域和任务特定模型。特别是在胸部X光(CXR)报告生成任务(MIMIC-CXR数据集)上,Med-PaLM M以超过8%的优势刷新了临床效能常用指标(micro-F1)的SOTA记录。在Multi Med Bench的医学视觉问答任务之一(Slake-VQA [12])中,Med-PaLM M在BLEU-1和F1指标上较此前SOTA结果提升了超过$10%$。
We perform ablation studies to understand the importance of scale in our generalist multimodal biomedical models and observe significant benefits for tasks that require higher-level language capabilities, such as medical (visual) question answering. Preliminary experiments also suggest evidence of zero-shot generalization to novel medical concepts and tasks across model scales, and emergent capabilities [13] such as zero-shot multimodal medical reasoning. We further perform radiologist evaluation of AI-generated chest X-ray reports and observe encouraging results across model scales.
我们通过消融实验来理解规模对通用多模态生物医学模型的重要性,并观察到在需要更高层次语言能力的任务(如医学(视觉)问答)中,规模带来的显著优势。初步实验还表明,不同规模的模型对新医学概念和任务表现出零样本泛化能力,并涌现出零样本多模态医学推理等新兴能力 [13]。我们进一步对AI生成的胸部X光报告进行放射科医师评估,观察到不同模型规模下均取得令人鼓舞的结果。
Overall, these results demonstrate the potential of generalist biomedical AI systems for medicine. However, significant work remains in terms of large-scale biomedical data access for training such models, validating performance in real world applications, and understanding the safety implications. We outline these key limitations and directions of future research in our study. To summarize, our key contributions are as follows:
总体而言,这些结果展现了通用生物医学AI系统在医疗领域的潜力。然而,要训练此类模型仍面临大规模生物医学数据获取、实际应用性能验证以及安全性影响理解等方面的重大挑战。我们在研究中概述了这些关键限制和未来研究方向。总结而言,我们的主要贡献如下:
2 Related Work
2 相关工作
2.1 Foundation models, multi modality, and generalists
2.1 基础模型、多模态与通用模型
The emergence of the foundation model paradigm [5] has had widespread impact across a variety of applications in language [8], vision [15], and other modalities [16]. While the idea of transfer learning [17, 18] using the weights of pretrained models has existed for decades [19–22], a shift has come about due to the scale of data and compute used for pre training such models [23]. The notion of a foundation model further indicates that the model can be adapted to a wide range of downstream tasks [5].
基础模型范式 [5] 的出现对语言 [8]、视觉 [15] 和其他模态 [16] 的多种应用产生了广泛影响。尽管利用预训练模型权重进行迁移学习 [17, 18] 的想法已存在数十年 [19–22],但由于此类模型预训练所采用的数据和计算规模 [23],这一领域发生了转变。基础模型的概念进一步表明,该模型可适配于广泛的下游任务 [5]。
Within the foundation model paradigm, multi modality [24] has also had a variety of important impacts – in the datasets [25], in the inter-modality supervision [26], and in the generality and unification of task specification [27, 28]. For example, language has specifically been an important enabler of foundation models in other modalities [11, 29]. Visual foundation models such as CLIP [30] are made possible by training on language-labeled visual datasets [25, 31], which are easier to collect from large-scale internet data than classification datasets with pre-determined class labels (i.e., ImageNet [32]). The benefits of joint languageand-vision supervision has also been noteworthy in generative modeling of images [33], where text-to-image generative modeling has been notably more successful at producing high-fidelity image generation [34] than purely un conditioned generative image modeling [35]. Further, the flexibility of language also enables a wide range of task specifications all via one unified output space [36] – it is possible to phrase tasks traditionally addressed by different output spaces, such as object detection and object classification, all jointly via the output space of language [37]. Med-PaLM M additionally benefits from the generality of multi modality, both via a model [10] pretrained on large vision-language datasets [11], and also by further biomedical domain finetuning through a unified generative language output space.
在基础模型(foundation model)范式下,多模态[24]也产生了多方面重要影响——体现在数据集[25]、跨模态监督[26]以及任务描述的通用性与统一性上[27, 28]。例如,语言特别成为其他模态中基础模型的重要赋能因素[11, 29]。视觉基础模型如CLIP[30]的实现,正是通过训练语言标注的视觉数据集[25, 31]完成的,这类数据比带有预定义类别标签的分类数据集(如ImageNet[32])更易于从大规模互联网数据中收集。语言-视觉联合监督的优势在图像生成建模[33]中也尤为显著,其中文本到图像的生成建模[34]在生成高保真图像方面明显优于纯无条件的图像生成建模[35]。此外,语言的灵活性还支持通过统一的输出空间[36]实现广泛的任务描述——传统上需要不同输出空间处理的任务(如目标检测和分类)都能通过语言的输出空间[37]统一表述。Med-PaLM M进一步受益于多模态的通用性,既通过基于大规模视觉-语言数据集[11]预训练的模型[10],也借助统一的生成式语言输出空间进行生物医学领域的微调。
A related notion to that of a foundation model is that of a generalist model – the same model with the same set of weights, without finetuning, can excel at a wide variety of tasks. A single multitask [17] model which can address many tasks has been of long standing interest [38, 39], including for example in the reinforcement learning community [40]. Language-only models such as GPT-3 [6] and PaLM [8] simultaneously excel at many tasks using only prompting and in-context learning. Recent work has also explored generalist models capable not only of performing many tasks, but also of processing many modalities [41]. For example, the capabilities of Gato [42] span language, vision, and agent policy learning. PaLM-E [10] further shows that it is possible to obtain a single generalist model which excels at language-only tasks, vision-language tasks, and embodied vision-language tasks. Med-PaLM M is specifically a generalist model designed for the biomedical domain, built by finetuning and aligning the PaLM-E generalist model.
与基础模型 (foundation model) 相关的概念是通用模型 (generalist model) —— 同一个模型使用同一组权重参数,无需微调 (finetuning) 即可在多种任务中表现出色。长期以来,能够处理多任务的单一模型 [17] 一直备受关注 [38, 39],例如强化学习领域 [40]。纯语言模型如 GPT-3 [6] 和 PaLM [8] 仅通过提示 (prompting) 和上下文学习 (in-context learning) 就能在多项任务中同时取得优异表现。近期研究还探索了不仅能执行多任务、还能处理多模态 [41] 的通用模型。例如 Gato [42] 的能力涵盖语言、视觉和智能体策略学习。PaLM-E [10] 进一步证明,可以获得一个在纯语言任务、视觉语言任务和具身视觉语言任务中都表现优异的单一通用模型。Med-PaLM M 是专为生物医学领域设计的通用模型,通过对 PaLM-E 通用模型进行微调和对齐构建而成。
2.2 Multimodal foundation models in bio medicine
2.2 生物医学中的多模态基础模型
Given the potential, there has been significant interest in multimodal foundation models for different biomedical applications. Moor et al. [43] discuss the notion of generalist medical AI, albeit without implementation or empirical results. Theodoris et al. [44] introduce Geneformer, a transformer [45] based model pretrained on a corpus of about 30 million single-cell transcript ome s to enable context-specific predictions in low data network biology applications. BiomedGPT [46] is a multi-task biomedical foundation model pretrained on a diverse source of medical images, medical literature, and clinical notes using a combination of language model (LM) and masked image infilling objectives. However, all these efforts are pretrained models and as such they require further task-specific data and finetuning to enable downstream applications. In contrast, Med-PaLM M is directly trained to jointly solve many biomedical tasks at the same time without requiring any further finetuning or model parameter updates. LLaVA-Med [47] is perhaps most similar to our effort. The authors use PubMed and GPT-4 [48] to curate a multimodal instruction following dataset and finetune a LLaVA model with it. However, the experiments are limited to three medical visual question answering datasets and qualitative examples of conversations conditioned on a medical image. In contrast, our work is more comprehensive, spanning multiple modalities including medical imaging, clinical text, and genomics with 14 diverse tasks and expert evaluation of model outputs.
鉴于其潜力,多模态基础模型在不同生物医学应用领域引起了极大兴趣。Moor等人[43]探讨了通用医疗AI的概念,但未提供具体实现或实证结果。Theodoris团队[44]提出了Geneformer——一种基于Transformer[45]的模型,该模型在约3000万个单细胞转录组数据集上进行了预训练,可用于低数据网络生物学应用中的上下文特异性预测。BiomedGPT[46]则是通过结合语言模型(LM)与掩码图像填充目标,在多样化医学图像、文献及临床记录上进行预训练的多任务生物医学基础模型。然而这些研究均为预训练模型,需要额外任务数据与微调才能支持下游应用。相比之下,Med-PaLM M直接通过联合训练同步解决多种生物医学任务,无需任何微调或参数更新。LLaVA-Med[47]可能与我们的工作最为相似,作者利用PubMed和GPT-4[48]构建了多模态指令跟随数据集并微调LLaVA模型,但其实验仅涵盖三个医学视觉问答数据集及基于医学图像的对话定性示例。而本研究覆盖医学影像、临床文本和基因组学等多模态领域,包含14项多样化任务及专家对模型输出的评估,具有更全面的研究维度。
2.3 Multimodal medical AI benchmarks
2.3 多模态医疗AI基准测试
To the best of our knowledge, there have been limited attempts to curate benchmarks for training and evaluating generalist biomedical AI models. Perhaps the work closest in spirit is BenchMD [49]. The benchmark spans 19 publicly available datasets and 7 medical modalities, including 1D sensor data, 2D images, and 3D volumetric scans. However, their tasks are primarily focused on classification whereas our benchmark also includes generative tasks such as medical (visual) question answering, radiology report generation and sum mari z ation. Furthermore, there is currently no implementation of a generalist biomedical AI system that can competently handle all these tasks simultaneously.
据我们所知,目前针对训练和评估通用生物医学AI模型构建基准的工作还十分有限。与此理念最接近的可能是BenchMD[49],该基准涵盖19个公开数据集和7种医学模态,包括1D传感器数据、2D图像和3D体积扫描。但他们的任务主要集中于分类,而我们的基准还包含生成式任务,如医疗(视觉)问答、放射学报告生成和摘要撰写。此外,目前尚未出现能同时胜任所有这些任务的通用生物医学AI系统实现。
3 Multi Med Bench: A Benchmark for Generalist Biomedical AI
3 Multi Med Bench: 通用生物医学AI基准测试
We next describe Multi Med Bench, a benchmark we curated to enable the development and evaluation of generalist biomedical AI. Multi Med Bench is a multi-task, multimodal benchmark comprising 12 de-identified open source datasets and 14 individual tasks. It measures the capability of a general-purpose biomedical AI to perform a variety of clinically-relevant tasks. The benchmark covers a wide range of data sources including medical questions, radiology reports, pathology, dermatology, chest X-ray, mammography, and genomics. Tasks in Multi Med Bench vary across the following axes:
接下来我们介绍Multi Med Bench,这是我们为促进通用生物医学AI(Artificial Intelligence)的开发和评估而精心策划的基准测试。Multi Med Bench是一个多任务、多模态的基准测试,包含12个去标识化的开源数据集和14个独立任务。它衡量通用生物医学AI执行各种临床相关任务的能力。该基准测试涵盖广泛的数据来源,包括医学问题、放射学报告、病理学、皮肤病学、胸部X光、乳腺X线摄影和基因组学。Multi Med Bench中的任务在以下维度上有所不同:
• Task type: question answering, report generation and sum mari z ation, visual question answering, medical image classification, and genomic variant calling. • Modality: text, radiology (CT, MRI, and X-ray), pathology, dermatology, mammography, and genomics. • Output format: open-ended generation for all tasks including classification.
- 任务类型:问答、报告生成与总结、视觉问答、医学图像分类和基因组变异识别。
- 模态:文本、放射学(CT、MRI和X光)、病理学、皮肤病学、乳腺X光摄影和基因组学。
- 输出格式:所有任务(包括分类)均采用开放式生成。
Table 1 | Multi Med Bench overview. Summary of Multi Med Bench, the benchmark we introduce for the development and evaluation of Med-PaLM M. Multi Med Bench consists of 14 individual tasks across 5 task types and 12 datasets spanning 7 biomedical data modalities. In total, the benchmark contains over 1 million samples.
| Task Type | Modality | Dataset | Description |
| Question Answering | Text | MedQA | US medical licensing exam-style, multiple-choice |
| MedMCQA | Indian medical entrance exams, multiple-choice | ||
| PubMedQA | Biomedical literature questions, multiple-choice | ||
| Report Summarization | Radiology | MIMIC-III | Summarizing findings in radiology reports |
| Visual Question Answering | Radiology | VQA-RAD | Close/open-ended VQA on radiology images |
| Slake-VQA | English-Chinese bilingual VQA on radiology images | ||
| Report Generation | Pathology Chest X-ray | Path-VQA MIMIC-CXR | Close/open-ended VQA on pathology images |
| Chest X-ray | MIMIC-CXR | Chest X-ray report generation | |
| Dermatology | PAD-UFES-20 | Binary classification of chest X-ray abnormalities 6-class skin lesion image classification | |
| Medical Image Classification | VinDr-Mammo | 5-classbreast-level BI-RADS classification | |
| Mammography | CBIS-DDSM | 3-class lesion-level classification (mass) | |
| CBIS-DDSM | 3-class lesion-level classification (calcification) | ||
| Genomics | PrecisionFDA | Genomic variant calling as 3-class image classification |
表 1 | Multi Med Bench概览。我们为Med-PaLM M的开发和评估引入的基准测试Multi Med Bench总结。该基准包含5种任务类型、12个数据集、7种生物医学数据模态的14项独立任务,总计超过100万个样本。
| 任务类型 | 模态 | 数据集 | 描述 |
|---|---|---|---|
| * * 问答* * | 文本 | MedQA | 美国医师执照考试风格多选题 |
| MedMCQA | 印度医学入学考试多选题 | ||
| PubMedQA | 生物医学文献多选题 | ||
| * * 报告摘要* * | 放射学 | MIMIC-III | 放射学报告发现总结 |
| * * 视觉问答* * | 放射学 | VQA-RAD | 放射影像封闭/开放式视觉问答 |
| Slake-VQA | 放射影像中英双语视觉问答 | ||
| * * 报告生成* * | 病理学胸部X光 | Path-VQA MIMIC-CXR | 病理影像封闭/开放式视觉问答 |
| 胸部X光 | MIMIC-CXR | 胸部X光报告生成 | |
| 皮肤病学 | PAD-UFES-20 | 胸部X光异常二分类6类皮肤病变图像分类 | |
| * * 医学图像分类* * | VinDr-Mammo | 5类乳腺BI-RADS分级 | |
| 乳腺X光 | CBIS-DDSM | 3类病灶级别分类(肿块) | |
| CBIS-DDSM | 3类病灶级别分类(钙化) | ||
| 基因组学 | PrecisionFDA | 基因组变异检测转为3类图像分类 |
Language-only tasks consist of medical question answering, including three of the MultiMedQA tasks used in Singhal et al. [9], and radiology report sum mari z ation. They were selected to assess a model’s ability to comprehend, recall, and manipulate medical knowledge. Multimodal tasks include medical visual question answering (VQA), medical image classification, chest X-ray report generation, and genomic variant calling, which are well-suited to evaluate both the visual understanding and multimodal reasoning capabilities of these models. Table 1 includes an overview of the datasets and tasks in Multi Med Bench - in total, the benchmark contains over 1 million samples. For detailed descriptions of individual datasets and tasks, see Section A.1.
纯语言任务包括医疗问答(涵盖Singhal等人[9]使用的三项MultiMedQA任务)和放射学报告摘要。这些任务旨在评估模型对医学知识的理解、记忆和运用能力。多模态任务则包含医疗视觉问答(VQA)、医学图像分类、胸部X光报告生成以及基因组变异识别,这些任务能有效评估模型的视觉理解与多模态推理能力。表1汇总了MultiMedBench中的数据集与任务概况——该基准测试共包含超过100万份样本。各数据集及任务的详细说明参见附录A.1节。
4 Med-PaLM M: A Proof of Concept for Generalist Biomedical AI
4 Med-PaLM M:通用生物医学AI的概念验证
In this section, we detail the methods underpinning the development of the Med-PaLM M model. We first review preliminaries of the pretrained models in Section 4.1 from which Med-PaLM M inherits, then discuss the datasets and training details involved in the finetuning and specialization of the model to the biomedical domain Section 4.2.
在本节中,我们将详细阐述支撑Med-PaLM M模型开发的方法。首先在第4.1节回顾Med-PaLM M所继承的预训练模型基础知识,随后在第4.2节讨论该模型在生物医学领域微调与专业化过程中涉及的数据集及训练细节。
4.1 Model preliminaries
4.1 模型基础
Note that Med-PaLM M inherits not only the architectures of these pretrained models, but also the general domain knowledge encoded in their model parameters.
需要注意的是,Med-PaLM M不仅继承了这些预训练模型的架构,还继承了编码在其模型参数中的通用领域知识。
Pathways Language Model (PaLM) introduced by Chowdhery et al. [8] is a densely-connected decoderonly Transformer [45] based large language model (LLM) trained using Pathways [50], a large-scale ML accelerator orchestration system that enables highly efficient training across TPU pods. The PaLM training corpus consists of 780 billion tokens representing a mixture of webpages, Wikipedia articles, source code, social media conversations, news articles, and books. PaLM models were trained at sizes of 8, 62, and 540 billion parameters, and all three PaLM model variants are trained for one epoch of the training data. At the time of its announcement, PaLM 540B achieved breakthrough performance, outperforming finetuned state-of-the-art models on a suite of multi-step reasoning tasks and exceeding average human performance on BIG-bench [51].
Chowdhery等人[8]提出的Pathways语言模型(PaLM)是基于密集连接纯解码器Transformer[45]的大语言模型(LLM),采用Pathways[50]系统训练。该大规模机器学习加速器编排系统能高效协调TPU pod集群进行训练。PaLM训练语料包含7800亿token,涵盖网页、维基百科、源代码、社交媒体对话、新闻文章和书籍。PaLM推出了80亿、620亿和5400亿参数三个版本,所有变体均在训练数据上训练一个完整周期。发布时,5400亿参数的PaLM在多项多步推理任务上超越精调的最先进模型,并在BIG-bench[51]基准上首次突破人类平均水平。
Vision Transformer (ViT) introduced by Do sov it ski y et al. [52] extends the Transformer [45] architecture to visual data such as images and videos. In this work, we consider two ViT pre-trained models as vision encoders, the 4 billion (4B) parameters model from Chen et al. [11] and the 22 billion (22B) parameters model from Dehghani et al. [15]. Both of these models were pretrained via supervised learning on a large classification dataset [53, 54] of approximately 4 billion images.
视觉Transformer (ViT) 由Dosovitskiy等人[52]提出,将Transformer[45]架构扩展至图像和视频等视觉数据。本文采用两个预训练的ViT模型作为视觉编码器:Chen等人[11]提出的40亿(4B)参数模型,以及Dehghani等人[15]提出的220亿(22B)参数模型。这两个模型均通过监督学习在包含约40亿图像的大型分类数据集[53,54]上完成预训练。
PaLM $\mathbf{E}$ introduced by Driess et al. [10] is a multimodal language model that can process sequences of multimodal inputs including text, vision, and sensor signals. The primary PaLM-E model uses pretrained PaLM and ViT, and was initially developed for embodied robotics applications but demonstrated strong performance on multiple vision language benchmarks such as OK-VQA [55] and VQA v2 [56]. Furthermore, PaLM-E offers the flexibility to interleave images, text and sensor signals in a single prompt, enabling the model to make predictions with a fully multimodal context. PaLM-E also exhibits a wide array of capabilities including zero-shot multimodal chain-of-thought (CoT) reasoning, and few-shot in-context learning. We therefore leverage the PaLM-E model as the base architecture for Med-PaLM M.
PaLM $\mathbf{E}$ 由 Driess 等人 [10] 提出,是一种多模态语言模型,能够处理包括文本、视觉和传感器信号在内的多模态输入序列。其主要模型基于预训练的 PaLM 和 ViT,最初为具身机器人应用开发,但在 OK-VQA [55] 和 VQA v2 [56] 等多个视觉语言基准测试中表现出色。此外,PaLM-E 支持在单个提示中灵活穿插图像、文本和传感器信号,使模型能够基于完整多模态上下文进行预测。该模型还具备零样本多模态思维链 (CoT) 推理和少样本上下文学习等广泛能力。因此,我们采用 PaLM-E 作为 Med-PaLM M 的基础架构。
We consider three different combinations of LLM and vision encoders in our study - PaLM 8B with ViT 4B (PaLM-E 12B), PaLM 62B with ViT 22B (PaLM-E 84B) and PaLM 540B with ViT 22B (PaLM-E 562B). All models were pretrained on diverse vision-language datasets in addition to tasks across multiple robot embodiment s as described in Driess et al. [10].
我们在研究中考虑了三种不同的大语言模型 (LLM) 和视觉编码器组合:ViT 4B 搭配 PaLM 8B (PaLM-E 12B)、ViT 22B 搭配 PaLM 62B (PaLM-E 84B) 以及 ViT 22B 搭配 PaLM 540B (PaLM-E 562B)。所有模型均在多样化视觉-语言数据集上进行预训练,并如 Driess 等人 [10] 所述,完成了跨多机器人实体形态的任务训练。
4.2 Putting it all together: Med-PaLM M
4.2 整体整合:Med-PaLM M
Med-PaLM M is developed by finetuning and aligning the PaLM-E model to the biomedical domain using Multi Med Bench. The following summarizes important methodological details underlying the development of the model.
Med-PaLM M是通过使用Multi Med Bench对PaLM-E模型进行微调和对齐到生物医学领域而开发的。以下总结了模型开发过程中的重要方法细节。
Dataset and preprocessing We resized all the images in Multi Med Bench to $224\times224\times3$ , while preserving the original aspect ratio with padding if needed. The gray-scale images were converted to 3-channel images by stacking up the same image along the channel dimension. Task-specific prepossessing methods such as class balancing and image data augmentation are described in detail for each task in Section A.1.
数据集与预处理
我们将Multi Med Bench中的所有图像尺寸调整为$224\times224\times3$,必要时通过填充保持原始宽高比。灰度图像通过沿通道维度堆叠相同图像转换为3通道图像。针对特定任务的预处理方法(如类别平衡和图像数据增强)将在附录A.1节中按任务详细说明。
Instruction task prompting and one-shot exemplar Our goal is to train a generalist biomedical AI model to perform multiple tasks with multimodal inputs using a unified model architecture and a single set of model parameters. To this end, we trained the model with a mixture of distinct tasks simultaneously via instruction tuning [57]. Specifically, we provided the model with task-specific instructions to prompt the model to perform different types of tasks in a unified generative framework. The task prompt consists of an instruction, relevant context information, and a question. For example, as shown in Figure 2, in the chest X-ray report generation task, we included the reason for the study and the image orientation information as additional context information for the model to condition its prediction on. Similarly, for the dermatology classification task, we provided the patient clinical history associated with the skin lesion image. We formulated all classification tasks as multiple choice questions where all possible class labels are provided as individual answer options and the model was prompted to generate the most likely answer as the target output. For other generative tasks such as visual question answering and report generation and sum mari z ation, the model was finetuned on the target response.
指令任务提示与单样本示例
我们的目标是训练一个通用的生物医学AI模型,使其能够使用统一的模型架构和单一参数集处理多模态输入下的多重任务。为此,我们通过指令调优[57]同时训练模型完成不同任务。具体而言,我们为模型提供任务特定的指令,引导其在统一的生成框架下执行各类任务。任务提示包含指令、相关上下文信息及问题。如图2所示,在胸部X光报告生成任务中,我们纳入检查原因和图像方位信息作为额外上下文,供模型预测时参考。同理,对于皮肤病分类任务,我们提供了与皮损图像相关的患者临床病史。所有分类任务均被构造成多选题形式,所有可能的类别标签作为独立选项给出,模型被要求生成最可能的答案作为目标输出。对于视觉问答、报告生成与摘要等其他生成任务,模型则针对目标响应进行微调。
In order to enable the model to better follow instructions, for the majority of tasks (see Table A.1), we added a text-only “one-shot exemplar” to the task prompt to condition the language model’s prediction. The one-shot exemplar helps prompt the model with a partial input-output pair. Importantly, for multimodal tasks, we replaced the actual image in the exemplar with a dummy text placeholder (with the text string “ $<$ <img $>$ ”): this (i) preserves training compute efficiency for single-image training, and also (ii) bypasses potential interference from cross-attention between a given text token and image tokens from multiple images [28]. Our results show that this scheme is effective in prompting the model to generate the desired format of responses as detailed in Section 6.
为了让模型更好地遵循指令,对于大多数任务(见表 A.1),我们在任务提示中添加了纯文本的"单样本示例"(one-shot exemplar)来调节语言模型的预测。单样本示例通过部分输入-输出对来引导模型。值得注意的是,对于多模态任务,我们将示例中的实际图像替换为虚拟文本占位符(文本字符串"$<$<img$>$"):这(i)保持了单图像训练的计算效率,(ii)避免了给定文本token与多张图像token之间交叉注意力可能产生的干扰[28]。结果显示,该方案能有效引导模型生成预期格式的响应,详见第6节。
Model training We finetuned the pretrained 12B, 84B, and 562B parameter variants of PaLM-E on Multi Med Bench tasks with mixture ratios denoted in Table A.1. These mixture ratios were empirically determined such that they are approximately proportional to the number of training samples in each dataset and ensuring at least one sample from each task is present in one batch. We performed an end-to-end
模型训练
我们在Multi Med Bench任务上对预训练的12B、84B和562B参数版本的PaLM-E进行了微调,混合比例如表A.1所示。这些混合比例是根据经验确定的,使其大致与每个数据集中的训练样本数量成比例,并确保每个任务至少有一个样本出现在一个批次中。我们进行了端到端的...
Instructions: You are a helpful radiology assistant. Describe what lines, tubes, and devices are present and each of their locations. Describe if pneumothorax is present; if present, describe size on each side. Describe if pleural eŦusion is present; if present, describe amount on each side. Describe if lung opacity (at elect as is, ůbrosis, consolidation, inůltrate, lung mass, pneumonia, pulmonary edema) is present; if present, describe kinds and locations. Describe the cardiac silhoueŵe size. Describe the width and contours of the media st in um. Describe if hilar enlargement is present; if enlarged, describe side. Describe what fractures or other skeletal abnormalities are present.
指令:你是一位专业的放射学助手。描述存在的导管、管路及设备及其各自位置。描述是否存在气胸;若存在,说明每侧的气胸大小。描述是否存在胸腔积液;若存在,说明每侧的积液量。描述是否存在肺部阴影(如肺不张、纤维化、实变、浸润、肺部肿块、肺炎、肺水肿);若存在,说明类型及位置。描述心脏轮廓大小。描述纵隔的宽度及轮廓。描述是否存在肺门增大;若增大,说明具体侧别。描述存在的骨折或其他骨骼异常。


Given the PA view X-ray image . Reason for the study: History m with malaise pneumonia. Q: Describe the ůndings in the image following the instructions. A:
给定胸部正位X光片。检查原因:肺炎伴不适病史。问:请根据指示描述影像所见。答:
Instructions: You are a helpful dermatology assistant. The following are questions about skin lesions. Categorize the skin lesions into the most likely class given the patient history.
说明:你是一位专业的皮肤科辅助助手。以下是关于皮肤病变的问题。根据患者病史将皮肤病变归类到最可能的类别。

Figure 2 | Illustration of instruction task prompting with one-shot exemplar. (top) shows the task prompt for the chest X-ray report generation task. It consists of task-specific instructions, a text-only “one-shot exemplar” (omitting the corresponding image but preserving the target answer), and the actual question. The X-ray image is embedded and interleaved with textual context including view orientation and reason for the study in addition to the question. (bottom) shows the task prompt for the dermatology classification task. We formulate the skin lesion classification task as a multiple choice question answering task with all the class labels provided as individual answer options. Similar to the chest X-ray report generation task, skin lesion image tokens are interleaved with the patient clinical history as additional context to the question. The blue denotes the position in the prompt where the image tokens are embedded.
图 2 | 单样本示例的指令任务提示示意图。(top) 展示了胸部X光报告生成任务的任务提示。它包含任务特定指令、纯文本的"单样本示例"(省略对应图像但保留目标答案)以及实际问题。X光图像被嵌入,并与包含视图方向和检查原因等文本上下文交错排列。(bottom) 展示了皮肤病分类任务的任务提示。我们将皮肤病变分类任务设计为多选题形式,所有类别标签都作为独立选项提供。与胸部X光报告生成任务类似,皮肤病变图像token与患者临床病史交错排列,作为问题的附加上下文。蓝色表示提示中嵌入图像token的位置。
finetuning of the PaLM-E model with the entire set of model parameters updated during training. For multimodal tasks, image tokens were interleaved with text tokens to form multimodal context input to the PaLM-E model. The multimodal context input contains at most 1 image for all finetuning tasks. However, we note that Med-PaLM M is able to process inputs with multiple images during inference.
在训练期间更新全部模型参数以对PaLM-E模型进行微调。对于多模态任务,图像token与文本token交错排列,形成PaLM-E模型的多模态上下文输入。所有微调任务的多模态上下文输入最多包含1张图像。但我们注意到,Med-PaLM M在推理时能够处理包含多张图像的输入。
We used the Adafactor optimizer [58] with momentum of $\beta_ {1}=0.9$ , dropout rate of 0.1, and a constant learning rate schedule. We used different sets of hyper parameters in our finetuning experiments for different model sizes, which are further detailed in Table A.2.
我们使用了Adafactor优化器[58],其动量$\beta_ {1}=0.9$、丢弃率为0.1,并采用恒定学习率调度。针对不同规模的模型,我们在微调实验中使用了不同的超参数集,具体细节见表A.2。
The resulting model, Med-PaLM M (12B, 84B, and 562B), is adapted to the biomedical domain with the capability to encode and interpret multimodal inputs and perform tasks including medical (visual) question answering, radiology report generation and sum mari z ation, medical image classification, and genomic variant calling.
最终得到的模型 Med-PaLM M (12B、84B 和 562B) 针对生物医学领域进行了适配,具备编码和解释多模态输入的能力,并能执行包括医学(视觉)问答、放射学报告生成与摘要、医学图像分类以及基因组变异检测在内的任务。
5 Evaluation
5 评估
In this section, we describe the purpose, scope, and methods of experimental evaluations. Results are presented in Section 6. Evaluation experiments of Med-PaLM M were designed for the following purposes:
在本节中,我们描述了实验评估的目的、范围和方法。结果将在第6节中展示。Med-PaLM M的评估实验设计用于以下目的:
• Evaluate generalist capabilities We evaluated Med-PaLM M on all tasks in Multi Med Bench across model scales. We provide initial insights on the effect of scaling ViT and LLM components across different tasks. We compared performance to previous SOTA (including specialist single-task or single-modality methods) and a state-of-art generalist model (PaLM-E) without biomedical finetuning. • Explore novel emergent capabilities One hypothesized benefit of training a single flexible multimodal generalist AI system across diverse tasks is the emergence of novel capabilities arising from language enabled combinatorial generalization, such as to novel medical concepts and tasks. We explored this via qualitative and qualitative experiments.
• 评估通用能力
我们在Multi Med Bench的所有任务上评估了不同规模下的Med-PaLM M模型,针对ViT和LLM组件在不同任务中的缩放效应提供了初步分析。性能对比对象包括先前的最先进技术(含单任务/单模态专家方法)以及未经生物医学微调的前沿通用模型(PaLM E)。
• 探索新兴能力
训练单一灵活的多模态通用AI系统的一个假设优势是:通过语言实现的组合泛化可能催生新能力(如处理新型医学概念和任务)。我们通过定性与定量实验对此进行了探索。
• Measure radiology report generation quality Automatic natural language generation (NLG) metrics do not provide sufficient evaluation of the clinical applicability of AI-generated radiology reports. We therefore performed expert radiologist evaluation of AI-generated reports on the MIMIC-CXR dataset, including comparison to the radiologist-provided reference reports.
• 衡量放射学报告生成质量
自动自然语言生成(NLG)指标无法充分评估AI生成放射学报告的临床适用性。因此我们邀请放射科专家对MIMIC-CXR数据集中AI生成的报告进行评估,包括与放射科医生提供的参考报告进行对比。
5.1 Evaluation on Multi Med Bench
5.1 在 Multi Med Bench 上的评估
Med-PaLM M was simultaneously finetuned on a mixture of language-only and multimodal biomedical tasks in Multi Med Bench. We assessed the model’s in-distribution performance on these tasks by comparing to the corresponding SOTA results obtained from separate specialist models. Specifically, we used the same few-shot setup as in training for each task during evaluation. Task-specific metrics were computed on the test split of each task and compared to prior SOTA specialist AI systems. Note that for a small number of tasks described in Table 1, we were not able to find a sufficiently similar prior attempt for comparison.
Med-PaLM M 在 Multi Med Bench 中同时针对纯语言和多模态生物医学任务进行了微调。我们通过比较从独立专家模型获得的相应 SOTA (State-of-the-Art) 结果,评估了模型在这些任务上的分布内性能。具体而言,在评估过程中,我们为每项任务使用了与训练时相同的少样本设置。针对每项任务的测试集计算了特定任务指标,并与先前的 SOTA 专家 AI 系统进行了比较。请注意,对于表 1 中描述的少数任务,我们未能找到足够相似的先前尝试进行比较。
5.2 Evaluation of language enabled zero-shot generalization
5.2 语言驱动的零样本泛化能力评估
To probe Med-PaLM M’s ability to generalize to previously unseen medical concepts, we evaluate the model’s ability to predict the presence or absence of tuberculosis (TB) from chest X-ray images. We used the Montgomery County chest X-ray set (MC) for this purpose. The dataset contains 138 frontal chest X-rays, of which 80 are normal cases and 58 cases have manifestations of TB [59]. Each case also contains annotations on the abnormality seen in the lung. We note that Med-PaLM M has been trained on MIMIC-CXR dataset; however, it is not trained to explicitly predict the TB disease label.
为了探究Med-PaLM M对未见过医学概念的泛化能力,我们评估了该模型通过胸部X光图像预测结核病(TB)存在与否的能力。为此,我们使用了蒙哥马利县胸部X光数据集(MC)。该数据集包含138张正面胸部X光片,其中80例为正常病例,58例有结核病表现[59]。每个病例还包含肺部异常的标注。需要说明的是,Med-PaLM M虽在MIMIC-CXR数据集上训练过,但并未专门针对结核病标签预测进行训练。
We evaluated the accuracy across model scales by formulating this problem as a two-choice question answering task where the model was prompted (with a text-only one-shot exemplar) to generate a yes/no answer about the presence of TB in the input image.
我们通过将该问题构建为一个二选一问答任务来评估不同规模模型的准确性,其中模型被提示(仅使用文本的单样本示例)生成关于输入图像中是否存在结核病 (TB) 的是/否答案。
We further explored zero-shot chain-of-thought (CoT) multimodal medical reasoning ability of the model by prompting with a text-only exemplar (without the corresponding image) and prompting the model to generate the class prediction and an accompanying report describing the image findings. We note that while we did prompt the model with a single text-only input-output pair, we omitted the image (used a dummy text placeholder instead) and the text exemplar was hand-crafted rather than drawn from the training set. Hence, this approach can be considered zero-shot rather than one-shot.
我们进一步探索了模型的零样本思维链(CoT)多模态医学推理能力,方法是通过纯文本示例(不含对应图像)提示模型,要求其生成类别预测并附带描述图像发现的报告。需要注意的是,虽然我们确实使用了单个纯文本输入-输出对作为提示,但省略了图像(改用虚拟文本占位符),且文本示例是人工构建而非来自训练集。因此,该方法应被视为零样本而非单样本。
In order to assess Med-PaLM M’s ability to generalize to novel task scenarios, we evaluated the model performance on two-view chest X-ray report generation - this is a novel task given the model was trained to generate reports only from a single-view chest X-ray.
为了评估 Med-PaLM M 在新任务场景中的泛化能力,我们在双视角胸部X光报告生成任务上测试了模型性能。需要指出的是,该模型仅接受过单视角胸部X光报告生成的训练,因此这项任务对其具有新颖性。
Finally, we also probed for evidence of positive task transfer as a result of jointly training a single generalist model to solve many different biomedical tasks. To this end, we performed an ablation study where we trained a Med-PaLM M 84B variant by excluding the MIMIC-CXR classification tasks from the task mixture. We compared this model variant to the Med-PaLM M 84B variant trained on the complete Multi Med Bench mixture on the chest X-ray report generation task with the expectation of improved performance in the latter.
最后,我们还探究了通过联合训练单一通用模型来解决多种不同生物医学任务时是否存在正向任务迁移的证据。为此,我们进行了消融实验:通过从任务混合集中排除MIMIC-CXR分类任务来训练一个Med-PaLM M 84B变体模型。我们将该变体模型与在完整Multi Med Bench混合集上训练的Med-PaLM M 84B变体模型进行胸片报告生成任务的性能对比,预期后者会表现出更好的性能。
5.3 Clinician evaluation of radiology report generation
5.3 临床医生对放射学报告生成的评估
To further assess the quality and clinical applicability of chest X-ray reports generated by Med-PaLM M and understand the effect of model scaling, we conducted a human evaluation using the MIMIC-CXR dataset. The evaluation was performed by four qualified thoracic radiologists based in India.
为了进一步评估Med-PaLM M生成的胸部X光报告的质量和临床适用性,并理解模型规模的影响,我们使用MIMIC-CXR数据集进行了人工评估。该评估由四位来自印度的合格胸科放射科医生完成。
Dataset The evaluation set consisted of 246 cases selected from the MIMIC-CXR test split. To match the expected input format of Med-PaLM M, we selected a single image from each study. We excluded studies that had ground truth reports mentioning multiple X-ray views or past examinations of the same patient.
数据集
评估集包含从MIMIC-CXR测试集中选取的246个病例。为匹配Med-PaLM M的预期输入格式,我们从每项研究中选取单张图像。我们排除了真实报告中提及多张X光视图或同一患者历史检查的研究。
Procedure We conducted two complementary human evaluations: (1) side-by-side evaluation where raters compared multiple alternative report findings and ranked them based on their overall quality, and (2)
流程
我们进行了两项互补的人工评估:(1) 并行评估,评分者比较多个备选报告结果并根据整体质量进行排序;(2)
independent evaluation where raters assessed the quality of individual report findings. Prior to performing the final evaluation, we iterated upon the instructions for the raters and calibrated their grades using a pilot set of 25 cases that were distinct from the evaluation set. Side-by-side evaluation was performed for all 246 cases, where each case was rated by a single radiologist randomly selected from a pool of four. For independent evaluation, each of the four radiologists independently annotated findings generated by three Med-PaLM M model variants (12B, 84B, and 562B) for every case in the evaluation set. Radiologists were blind to the source of the report findings for all evaluation tasks, and the reports were presented in a randomized order.
独立评估中,评分者对单个报告结果的质量进行评判。在开展最终评估前,我们通过25例独立于评估集的试点案例,对评分指南进行迭代优化并对评分标准进行校准。所有246例均采用并行评估方式,每例由四位放射科医生中随机指定的一位进行评分。在独立评估环节,四位放射科医生分别对评估集中每例病例的三个Med-PaLM M模型变体(12B/84B/562B)生成结果进行标注。所有评估任务中,放射科医生均不知晓报告结果的来源,且报告以随机顺序呈现。
Side-by-side evaluation The input to each side-by-side evaluation was a single chest X-ray, along with the “indication” section from the MIMIC-CXR study. Four alternative options for the “findings” section of the report were shown to raters as depicted in Figure A.3. The four alternative “findings” sections corresponded to the dataset reference report’s findings, and findings generated by three Med-PaLM M model variants (12B, 84B, 562B). Raters were asked to rank the four alternative findings based on their overall quality using their best clinical judgement.
并行评估
每次并行评估的输入是一张胸部X光片,以及来自MIMIC-CXR研究的"适应症(indication)"部分。如图A.3所示,评估者会看到报告"结果(findings)"部分的四种备选方案。这四种备选"结果"分别对应数据集参考报告的结果,以及三个Med-PaLM M模型变体(12B、84B、562B)生成的结果。评估者被要求根据其最佳临床判断,对这四种备选结果的整体质量进行排序。
Independent evaluation For independent evaluation, raters were also presented with a single chest X-ray, along with the indication and reference report’s findings from the MIMIC-CXR study (marked explicitly as such), but this time only a single findings paragraph generated by Med-PaLM M as shown in Figure A.4. Raters were asked to assess the quality of the Med-PaLM M generated findings in the presence of the reference inputs provided and their own judgement of the chest X-ray image. The rating schema proposed in Yu et al. [60] served as inspiration for our evaluation task design.
独立评估
在独立评估中,评审员会看到一张胸部X光片、检查指征以及来自MIMIC-CXR研究的参考报告结果(明确标注来源),但这次仅提供如图A.4所示的Med-PaLM M生成的单个结果段落。评审员需根据提供的参考输入及其对胸部X光图像的独立判断,评估Med-PaLM M生成结果的质量。我们的评估任务设计参考了Yu等人[60]提出的评分框架。
First, raters assessed whether the quality and view of the provided image were sufficient to perform the evaluation task fully. Next, they annotated all passages in the model-generated findings that they disagreed with (errors), and all missing parts (omissions). Raters categorized each error passage by its type (no finding, incorrect finding location, incorrect severity, reference to non-existent view or prior study), assessed its clinical significance, and suggested alternative text to replace the selected passage. Likewise, for each omission, raters specified a passage that should have been included and determined if the omission had any clinical significance.
首先,评分者评估所提供图像的质量和视角是否足以完整执行评估任务。接着,他们标注了模型生成报告中所有不认同的段落(错误)以及遗漏的部分(缺失)。评分者按类型对每个错误段落进行分类(无发现、错误发现位置、错误严重程度、引用不存在的视角或先前研究),评估其临床意义,并建议替换所选段落的替代文本。同样,对于每个缺失部分,评分者指定了应包含的段落,并判断该缺失是否具有临床意义。
6 Results
6 结果
Here we present results across the three different evaluation setups introduced in Section 5.
在此我们展示第5节介绍的三种不同评估设置下的结果。
6.1 Med-PaLM M performs near or exceeding SOTA on all Multi Med Bench tasks
6.1 Med-PaLM M 在所有 Multi Med Bench 任务上的表现接近或超越 SOTA
Med-PaLM M performance versus baselines We compared Med-PaLM M with two baselines:
Med-PaLM M 性能对比基线
我们将 Med-PaLM M 与两个基线进行了比较:
Results are summarized in Table 2. Across Multi Med Bench tasks, Med-PaLM M’s best result (across three model sizes) exceeded prior SOTA results on 5 out of 12 tasks (for two tasks, we were unable to find a prior SOTA comparable to our setup) while being competitive on the rest. Notably, these results were achieved with a generalist model using the same set of model weights without any task-specific architecture customization or optimization.
结果总结在表2中。在Multi Med Bench的各项任务中,Med-PaLM M的最佳表现(涵盖三种模型规模)在12项任务中有5项超过了之前的SOTA结果(对于其中两项任务,我们未能找到与我们的设置可比的先前SOTA),同时在其他任务上保持竞争力。值得注意的是,这些成绩是由一个通用模型实现的,该模型使用同一组权重,未进行任何针对特定任务的架构定制或优化。
On medical question answering tasks, we compared against the SOTA Med-PaLM 2 results [61] and observed higher performance of Med-PaLM 2. However, when compared to the baseline PaLM model on which Med-PaLM M was built, Med-PaLM M outperformed the previous best PaLM results [9] by a large margin in the same few-shot setting on all three question answering datasets.
在医疗问答任务上,我们与当前最佳 (SOTA) 的 Med-PaLM 2 结果 [61] 进行了对比,观察到 Med-PaLM 2 表现更优。然而,与构建 Med-PaLM M 所用的基线 PaLM 模型相比,Med-PaLM M 在三个问答数据集上的相同少样本设置中,均大幅超越了此前 PaLM 的最佳结果 [9]。
Further, when compared to PaLM-E 84B as a generalist baseline without biomedical domain finetuning, MedPaLM M exhibited performance improvements on all 14 tasks often by a significant margin, demonstrating the importance of domain adaptation. Taken together, these results illustrate the strong capabilities of Med-PaLM M as a generalist biomedical AI model. We further describe the results in detail for each of the individual tasks in Section A.3.
此外,与未经生物医学领域微调的通用基线模型PaLM-E 84B相比,MedPaLM M在所有14项任务上均表现出性能提升,且优势往往显著,这证明了领域适应的重要性。总体而言,这些结果展现了Med-PaLM M作为通用生物医学AI模型的强大能力。我们将在章节A.3中详细描述各项任务的独立结果。
Table 2 | Performance comparison on Multi Med Bench. We compare Med-PaLM M with specialist SOTA models and a generalist model (PaLM-E 84B) without biomedical domain finetuning. Across all tasks, datasets and metrics combination in Multi Med Bench, we observe Med-PaLM M performance near or exceeding SOTA. Note that these results are achieved by Med-PaLM M with the same set of model weights without any task-specific customization.
| Task Type | Modality | Dataset | Metric | SOTA | PaLM-E | Med-PaLM M (Best) |
| (84B) | ||||||
| Question Answering | Text | MedQA | Accuracy | 86.50% [61] | 28.83% | 69.68% |
| MedMCQA | Accuracy | 72.30% [61] | 33.35% | 62.59% | ||
| PubMedQA | Accuracy | 81.80% [61] | 64.00% | 80.00% | ||
| Report Summarization | Radiology | MIMIC-III | ROUGE-L | 38.70% [62] | 3.30% | 32.03% |
| BLEU | 16.20% [62] | 0.34% | 15.36% | |||
| F1-RadGraph | 40.80% [62] | 8.00% | 34.71% | |||
| Visual Question Answering | Radiology | VQA-RAD | BLEU-1 | 71.03% [63] | 59.19% | 71.27% |
| F1 | N/A | 38.67% | 62.06% | |||
| Slake-VQA | BLEU-1 | 78.60% [64] | 52.65% | 92.7% | ||
| F1 | 78.10% [64] | 24.53% | 89.28% | |||
| Pathology | Path-VQA | BLEU-1 F1 | 70.30% [64] 58.40% [64] | 54.92% 29.68% | 72.27% 62.69% | |
| Report Generation | Chest X-ray | MIMIC-CXR | Micro-F1-14 | 44.20% [65] | 15.40% | 53.56% |
| Macro-F1-14 | 30.70% [65] | 10.11% | 39.83% | |||
| Micro-F1-5 | 56.70% [66] | 5.51% | 57.88% | |||
| Macro-F1-5 | N/A | 4.85% | 51.60% | |||
| F1-RadGraph | 24.40% [14] | 11.66% | 26.71% | |||
| BLEU-1 | 39.48% [65] | 19.86% | 32.31% | |||
| BLEU-4 | 13.30% [66] | 4.60% | 11.50% | |||
| ROUGE-L | 29.60% [67] | 16.53% | 27.49% | |||
| CIDEr-D | 49.50% [68] | 3.50% | 26.17% | |||
| MIMIC-CXR Macro-AUC | 81.27% [69] | 51.48% | 79.09% | |||
| (5 conditions) Dermatology PAD-UFES-20 | Macro-F1 | N/A N/A | 7.83% 63.37% | 41.57% | ||
| Macro-AUC | N/A | 1.38% | 97.27% 84.32% | |||
| VinDr-Mammo | Macro-F1 Macro-AUC | 64.50% [49] | 51.49% | 71.76% | ||
| Macro-F1 | N/A | 16.06% | 35.70% | |||
| CBIS-DDSM Mammography | Macro-AUC | N/A | 47.75% | 73.31% | ||
| Macro-F1 | N/A | 7.77% | 51.12% | |||
| (mass) CBIS-DDSM | 40.67% | |||||
| (calcification) | Macro-AUC Macro-F1 | N/A 70.71% [70] | 11.37% | 82.22% 67.86% | ||
| Genomics PrecisionFDA | Indel-F1 | 99.40% [71] | 53.01% | 97.04% | ||
| (Variant Calling) (Truth Challenge V2) | SNP-F1 | 99.70% [71] | 52.84% 99.35% | |||
表 2 | Multi Med Bench性能对比。我们将Med-PaLM M与专业领域的SOTA模型及未经生物医学领域微调的通用模型(PaLM-E 84B)进行对比。在Multi Med Bench所有任务、数据集和指标组合中,Med-PaLM M表现接近或超越SOTA。请注意,这些结果是Med-PaLM M使用同一组模型权重且未进行任何任务特定定制所取得的。
| 任务类型 | 模态 | 数据集 | 指标 | SOTA | PaLM-E (84B) | Med-PaLM M (最佳) |
|---|---|---|---|---|---|---|
| * * 问答* * | 文本 | MedQA | 准确率 | 86.50% [61] | 28.83% | 69.68% |
| MedMCQA | 准确率 | 72.30% [61] | 33.35% | 62.59% | ||
| PubMedQA | 准确率 | 81.80% [61] | 64.00% | 80.00% | ||
| * * 报告摘要* * | 放射学 | MIMIC-III | ROUGE-L | 38.70% [62] | 3.30% | 32.03% |
| BLEU | 16.20% [62] | 0.34% | 15.36% | |||
| F1-RadGraph | 40.80% [62] | 8.00% | 34.71% | |||
| * * 视觉问答* * | 放射学 | VQA-RAD | BLEU-1 | 71.03% [63] | 59.19% | 71.27% |
| F1 | N/A | 38.67% | 62.06% | |||
| Slake-VQA | BLEU-1 | 78.60% [64] | 52.65% | 92.7% | ||
| F1 | 78.10% [64] | 24.53% | 89.28% | |||
| 病理学 | Path-VQA | BLEU-1 F1 | 70.30% [64] 58.40% [64] | 54.92% 29.68% | 72.27% 62.69% | |
| * * 报告生成* * | 胸部X光 | MIMIC-CXR | Micro-F1-14 | 44.20% [65] | 15.40% | 53.56% |
| Macro-F1-14 | 30.70% [65] | 10.11% | 39.83% | |||
| Micro-F1-5 | 56.70% [66] | 5.51% | 57.88% | |||
| Macro-F1-5 | N/A | 4.85% | 51.60% | |||
| F1-RadGraph | 24.40% [14] | 11.66% | 26.71% | |||
| BLEU-1 | 39.48% [65] | 19.86% | 32.31% | |||
| BLEU-4 | 13.30% [66] | 4.60% | 11.50% | |||
| ROUGE-L | 29.60% [67] | 16.53% | 27.49% | |||
| CIDEr-D | 49.50% [68] | 3.50% | 26.17% | |||
| MIMIC-CXR (5种病症) | Macro-AUC | 81.27% [69] | 51.48% | 79.09% | ||
| 皮肤病学 | PAD-UFES-20 | Macro-F1 | N/A | 7.83% | 41.57% | |
| Macro-AUC | N/A | 63.37% | 97.27% | |||
| VinDr-Mammo | Macro-F1 | 64.50% [49] | 51.49% | 71.76% | ||
| Macro-AUC | N/A | 16.06% | 35.70% | |||
| CBIS-DDSM乳腺摄影 | Macro-AUC | N/A | 47.75% | 73.31% | ||
| Macro-F1 | N/A | 7.77% | 51.12% | |||
| CBIS-DDSM (肿块) | 40.67% | |||||
| CBIS-DDSM (钙化) | Macro-AUC | N/A | 11.37% | 82.22% | ||
| Macro-F1 | 70.71% [70] | 67.86% | ||||
| * * 基因组学* * | 变异检测 | PrecisionFDA (Truth Challenge V2) | Indel-F1 | 99.40% [71] | 53.01% | 97.04% |
| SNP-F1 | 99.70% [71] | 52.84% | 99.35% |
Med-PaLM M performance across model scales We summarize Med-PaLM M performance across model scales (12B, 84B, and 562B) in Table 3. The key observations are:
不同规模Med-PaLM M模型的性能表现
我们在表3中总结了Med-PaLM M在三种模型规模(12B、84B和562B)下的性能表现。主要观察结果包括:
• Language reasoning tasks benefit from scale For tasks that require language understanding and reasoning such as medical question answering, medical visual question answering and radiology report sum mari z ation, we see significant improvements as we scale up the model from 12B to 562B. • Multimodal tasks bottle necked by vision encoder performance For tasks such as mammography or dermatology image classification, where nuanced visual understanding is required but minimal language reasoning is needed (outputs are classification label tokens only), the performance improved from Med-PaLM M 12B to Med-PaLM 84B but plateaued for the 562B model, possibly because the vision encoder is not further scaled in that step (both the Med-PaLM M 84B and 562B models use the same 22B ViT as the vision encoder), thereby acting as a bottleneck to observing a scaling benefit. We note the possibility of additional con founders here such as the input image resolution.
- 语言推理任务受益于模型规模 在需要语言理解和推理的任务(如医学问答、医学视觉问答和放射学报告总结)中,当模型规模从120亿参数扩展到5620亿参数时,我们观察到显著性能提升。
- 视觉编码器性能制约多模态任务表现 对于需要精细视觉理解但仅需最低限度语言推理的任务(如乳腺X光或皮肤病图像分类,其输出仅为分类标签token),模型性能从Med-PaLM M 120亿提升到Med-PaLM 840亿后趋于稳定,在5620亿参数模型中未现进一步增益。这可能因为该阶段未同步扩展视觉编码器规模(Med-PaLM M 840亿和5620亿模型均采用相同的220亿参数ViT视觉编码器),从而成为观察扩展效益的瓶颈。我们注意到其他潜在影响因素的存在,例如输入图像分辨率等。
The scaling results on the chest X-ray report generation task are interesting (Table 3). While on the surface, the task seems to require complex language understanding and reasoning capabilities and would thus benefit from scaling the language model, we find the Med-PaLM M 84B model to be roughly on-par or slightly exceeding the 562B model on a majority of metrics, which may simply be due to fewer training steps used for the larger model. Another possibility for the diminishing return of increasing the size of language model is likely that the output space for chest X-ray report generation in the MIMIC-CXR dataset is fairly confined to a set of template sentences and limited number of conditions. This insight has motivated the use of retrieval based approaches as opposed to a fully generative approach for the chest X-ray report generation task on this dataset [72, 73]. Additionally, the larger 562B model has a tendency towards verbosity rather than the comparative brevity of the 84B model, and without further preference alignment in training, this may impact its metrics.
胸部X光报告生成任务的扩展结果很有趣(表3)。虽然表面上这项任务似乎需要复杂的语言理解和推理能力,因此会受益于大语言模型的扩展,但我们发现Med-PaLM M 84B模型在大多数指标上与562B模型大致相当或略胜一筹,这可能仅仅是由于较大模型使用的训练步骤较少。语言模型规模增加带来的收益递减的另一个可能原因是,MIMIC-CXR数据集中胸部X光报告生成的输出空间相当局限于一组模板句子和有限数量的条件。这一见解促使在该数据集的胸部X光报告生成任务中采用基于检索的方法,而不是完全生成式方法[72,73]。此外,较大的562B模型倾向于冗长,而不是84B模型的相对简洁,如果在训练中没有进一步的偏好对齐,这可能会影响其指标。
Table 3 | Performance of Med-PaLM M on Multi Med Bench across model scales. We summarize the performance of Med-PaLM M across three model scale variants 12B, 84B, 562B. All models were finetuned and evaluated on the same set of tasks in Multi Med Bench. We observe that scaling plays a key role in language-only tasks and multimodal tasks that require reasoning such as visual question answering. However, scaling has diminishing benefit for image classification and chest X-ray report generation task.
| Task Type | Modality | Dataset | Metric | Med-PaLM M | Med-PaLM M (84B) | Med-PaLM M (562B) |
| (12B) | ||||||
| Question Answering | Text | MedQA | Accuracy | 29.22% | 46.11% | 69.68% |
| MedMCQA | Accuracy | 32.20% | 47.60% | 62.59% | ||
| PubMedQA | Accuracy | 48.60% | 71.40% | 80.00% | ||
| MIMIC-III | ROUGE-L | 29.45% | 31.47% | 32.03% | ||
| Radiology | BLEU | 12.14% | 15.36% | 15.21% | ||
| F1-RadGraph | 31.43% | 33.96% | 34.71% | |||
| VQA-RAD | BLEU-1 | 64.02% | 69.38% | 71.27% | ||
| F1 | 50.66% | 59.90% | 62.06% | |||
| BLEU-1 Slake-VQA | 90.77% | 92.70% | 91.64% | |||
| Pathology | Path-VQA | F1 | 86.22% | 89.28% | 87.50% | |
| BLEU-1 | 68.97% 57.24% | 70.16% 59.51% | 72.27% 62.69% | |||
| F1 Micro-F1-14 | 51.41% | 53.56% | 51.60% | |||
| 37.31% | 39.83% | 37.81% | ||||
| Macro-F1-14 Micro-F1-5 | 56.54% | 57.88% | 56.28% | |||
| Macro-F1-5 | 50.57% | 51.60% | 49.86% | |||
| F1-RadGraph | 25.20% | 26.71% | 26.06% | |||
| MIMIC-CXR BLEU-1 | 30.90% | 32.31% | 31.73% | |||
| BLEU-4 | 10.43% | 11.31% | 11.50% | |||
| ROUGE-L | 26.16% | 27.29% | 27.49% | |||
| CIDEr-D | 23.43% | 26.17% | 25.27% | |||
| MIMIC-CXR Chest X-ray (5 conditions) | Macro-AUC | 76.67% | 78.35% | 79.09% | ||
| Macro-F1 | 38.33% | 36.83% | 41.57% | |||
| Dermatology | PAD-UFES-20 | Macro-AUC | 95.57% | 97.27% | 96.08% | |
| Macro-F1 | 78.42% | 84.32% | 77.03% | |||
| VinDr-Mammo | Macro-AUC 66.29% | 71.76% | 71.42% | |||
| Mammography | Macro-F1 | 29.81% | 35.70% | 33.90% | ||
| CBIS-DDSM | Macro-AUC | 70.11% | 73.09% 73.31% | |||
| (mass) | Macro-F1 | 47.23% | 49.98% | 51.12% | ||
| CBIS-DDSM | Macro-AUC | 81.40% | 82.22% | 80.90% | ||
| (calcification) | Macro-F1 | 67.86% | 63.81% | 63.03% | ||
| Genomics Variant Calling | Indel-F1 | 96.42% | 97.04% | 95.46% | ||
| SNP-F1 | 99.35% | 99.32% | 99.16% |
表 3 | Med-PaLM M 在 Multi Med Bench 不同模型规模下的性能表现。我们总结了 Med-PaLM M 在 12B、84B 和 562B 三种规模变体上的性能表现。所有模型都在 Multi Med Bench 的相同任务集上进行了微调和评估。我们观察到,模型规模对纯语言任务和需要推理的多模态任务(如视觉问答)起着关键作用。然而,对于图像分类和胸部 X 光报告生成任务,模型规模的收益会递减。
| 任务类型 | 模态 | 数据集 | 指标 | Med-PaLM M (12B) | Med-PaLM M (84B) | Med-PaLM M (562B) |
|---|---|---|---|---|---|---|
| 问答 | 文本 | MedQA | 准确率 | 29.22% | 46.11% | 69.68% |
| MedMCQA | 准确率 | 32.20% | 47.60% | 62.59% | ||
| PubMedQA | 准确率 | 48.60% | 71.40% | 80.00% | ||
| MIMIC-III | ROUGE-L | 29.45% | 31.47% | 32.03% | ||
| 放射学 | Radiology | BLEU | 12.14% | 15.36% | 15.21% | |
| F1-RadGraph | 31.43% | 33.96% | 34.71% | |||
| VQA-RAD | BLEU-1 | 64.02% | 69.38% | 71.27% | ||
| F1 | 50.66% | 59.90% | 62.06% | |||
| Slake-VQA | BLEU-1 | 90.77% | 92.70% | 91.64% | ||
| 病理学 | Path-VQA | F1 | 86.22% | 89.28% | 87.50% | |
| BLEU-1 | 68.97% 57.24% | 70.16% 59.51% | 72.27% 62.69% | |||
| F1 Micro-F1-14 | 51.41% | 53.56% | 51.60% | |||
| Macro-F1-14 | 37.31% | 39.83% | 37.81% | |||
| Micro-F1-5 | 56.54% | 57.88% | 56.28% | |||
| Macro-F1-5 | 50.57% | 51.60% | 49.86% | |||
| F1-RadGraph | 25.20% | 26.71% | 26.06% | |||
| MIMIC-CXR | BLEU-1 | 30.90% | 32.31% | 31.73% | ||
| BLEU-4 | 10.43% | 11.31% | 11.50% | |||
| ROUGE-L | 26.16% | 27.29% | 27.49% | |||
| CIDEr-D | 23.43% | 26.17% | 25.27% | |||
| MIMIC-CXR Chest X-ray (5 conditions) | Macro-AUC | 76.67% | 78.35% | 79.09% | ||
| Macro-F1 | 38.33% | 36.83% | 41.57% | |||
| 皮肤病学 | PAD-UFES-20 | Macro-AUC | 95.57% | 97.27% | 96.08% | |
| Macro-F1 | 78.42% | 84.32% | 77.03% | |||
| VinDr-Mammo | Macro-AUC | 66.29% | 71.76% | 71.42% | ||
| Macro-F1 | 29.81% | 35.70% | 33.90% | |||
| 乳腺摄影 | CBIS-DDSM (mass) | Macro-AUC | 70.11% | 73.09% 73.31% | ||
| Macro-F1 | 47.23% | 49.98% | 51.12% | |||
| CBIS-DDSM (calcification) | Macro-AUC | 81.40% | 82.22% | 80.90% | ||
| Macro-F1 | 67.86% | 63.81% | 63.03% | |||
| 基因组学变异检测 | Indel-F1 | 96.42% | 97.04% | 95.46% | ||
| SNP-F1 | 99.35% | 99.32% | 99.16% |
6.2 Med-PaLM M demonstrates zero-shot generalization to novel medical tasks and concepts
6.2 Med-PaLM M 展示了对新型医疗任务和概念的零样本 (Zero-shot) 泛化能力
Training a generalist biomedical AI system with language as a common grounding across different tasks allows the system to tackle new tasks by combining the knowledge it has learned for other tasks (i.e. combinatorial generalization). We highlight preliminary evidence which suggests Med-PaLM M can generalize to novel medical concepts and unseen tasks in a zero-shot fashion. We further observe zero-shot multimodal reasoning as an emergent capability [13] of Med-PaLM M. Finally, we demonstrate benefits from positive task transfer as a result of the model’s multi-task, multimodal training.
训练一个通用型生物医学AI系统,以语言作为跨任务统一基础,使系统能够通过组合已学知识应对新任务(即组合泛化)。我们展示了初步证据表明Med-PaLM M能以零样本方式泛化到新医学概念和未见任务。进一步观察到零样本多模态推理是Med-PaLM M的涌现能力[13]。最后,我们验证了该模型通过多任务多模态训练获得的正向任务迁移优势。
6.2.1 Evidence of generalization to novel medical concepts
6.2.1 对新医学概念的泛化能力证据
We probed the zero-shot generalization capability of Med-PaLM M for an unseen medical concept by evaluating its ability to detect tuberculosis (TB) abnormality from chest X-ray images in the Montgomery County (MC) dataset. As shown in Table 4, Med-PaLM M performed competitively compared to SOTA results obtained by a specialized ensemble model optimized for this dataset [74]. We observed similar performance across three model variants, consistent with findings on other medical image classification tasks in Multi Med Bench. Given the classification task was set up as an open-ended question answering task, we did not report the AUC metric which requires the normalized predicted probability of each possible class.
我们通过评估Med-PaLM M在蒙哥马利县(MC)数据集的胸部X光图像中检测结核病(TB)异常的能力,探究了其对未见过医学概念的零样本泛化能力。如表4所示,与针对该数据集优化的专用集成模型[74]获得的最先进(SOTA)结果相比,Med-PaLM M表现具有竞争力。我们在三个模型变体中观察到相似的性能,这与Multi Med Bench中其他医学图像分类任务的研究结果一致。由于该分类任务被设置为开放式问答任务,我们未报告需要每个可能类别的归一化预测概率的AUC指标。
Table 4 | Zero-shot classification performance of Med-PaLM M on the tuberculosis (TB) detection task. Med-PaLM M performs competitively to the SOTA model [74] finetuned on the Montgomery County TB dataset using model ensemble. Notably, Med-PaLM M achieves this result with a simple task prompt consisting of a single text-only exemplar (without task-specific image and hence zero-shot), in contrast to the specialist model that requires training on all the samples in the dataset.
| Model | # Training samples | Accuracy |
| SOTA 1 [74] | 138 | 92.60% |
| Med-PaLM M (12B) | 0 | 86.96% |
| Med-PaLM M (84B) | 0 | 82.60% |
| Med-PaLM M (562B) | 0 | 87.68% |
表 4 | Med-PaLM M 在结核病 (TB) 检测任务中的零样本分类性能。Med-PaLM M 的表现与在 Montgomery County TB 数据集上通过模型集成微调的 SOTA 模型 [74] 相当。值得注意的是,Med-PaLM M 仅通过一个纯文本示例的简单任务提示(不含任务特定图像,因此为零样本)就实现了这一结果,而专业模型需要在数据集中的所有样本上进行训练。
| 模型 | 训练样本数 | 准确率 |
|---|---|---|
| SOTA 1 [74] | 138 | 92.60% |
| Med-PaLM M (12B) | 0 | 86.96% |
| Med-PaLM M (84B) | 0 | 82.60% |
| Med-PaLM M (562B) | 0 | 87.68% |
6.2.2 Evidence of emergent zero-shot multimodal medical reasoning
6.2.2 涌现的零样本多模态医学推理证据
We also qualitatively explored the zero-shot chain-of-thought (CoT) capability of Med-PaLM M on the MC TB dataset. In contrast to the classification setup, we prompted the model with a text-only exemplar to generate a report describing the findings in a given image in addition to a yes/no classification prediction. In Figure 3, we present qualitative examples of zero-shot CoT reasoning from the Med-PaLM M 84B and 562B variants. In particular, both Med-PaLM M variants were able to identify the major TB related lesion in the correct location. However, according to expert radiologist review, there are still some omissions of findings and errors in the model generated report, suggesting room for improvement. It is noteworthy that Med-PaLM M 12B failed to generate a coherent visually conditioned response, which indicates that scaling of the language model plays a key role in the zero-shot CoT multimodal reasoning capability (i.e. this might be an emergent capability [13]).
我们还对Med-PaLM M在MC TB数据集上的零样本思维链(CoT)能力进行了定性探索。与分类设置不同,我们向模型提供了一个纯文本示例,要求其除了生成是/否分类预测外,还要生成描述给定图像中发现的报告。在图3中,我们展示了Med-PaLM M 84B和562B变体的零样本CoT推理定性示例。值得注意的是,两个Med-PaLM M变体都能在正确位置识别出与结核病相关的主要病变。然而,根据放射科专家评审,模型生成的报告中仍存在一些遗漏发现和错误,表明还有改进空间。需要指出的是,Med-PaLM M 12B未能生成连贯的视觉条件响应,这表明语言模型的规模在零样本CoT多模态推理能力中起着关键作用(即这可能是一种涌现能力[13])。
6.2.3 Evidence of generalization to novel tasks
6.2.3 对新任务的泛化能力证据
Although Med-PaLM M was only trained with single-view chest X-ray image inputs, we observed the capability of the model to generalize to a novel task setup with multi-view visual inputs. Specifically, on a subset of studies from MIMIC-CXR where each report is accompanied with both a frontal and a lateral view X-ray image. we observe that Med-PaLM M is able to attain zero-shot performance comparable to the single-view report generation task as detailed in Table 5. This ability is promising given medical imaging studies often benefit from the interpretation of prior historical studies in addition to the current instance for optimal performance.
尽管 Med-PaLM M 仅使用单视角胸部 X 射线图像输入进行训练,但我们观察到该模型能够泛化到具有多视角视觉输入的新任务设置。具体而言,在 MIMIC-CXR 的一个子集研究中,每份报告都配有正面和侧面 X 射线图像。我们观察到 Med-PaLM M 能够达到与单视角报告生成任务相当的零样本性能,详见表 5。考虑到医学影像研究通常需要结合当前实例和历史先例的解读以获得最佳性能,这一能力颇具前景。
6.2.4 Evidence of positive task transfer
6.2.4 正向任务迁移的证据
To demonstrate the positive task transfer arising from joint training across modalities and tasks, we performed an ablation study where we trained a Med-PaLM M 84B variant by excluding the MIMIC-CXR classification task from the task mixture and compared this model variant against Med-PaLM M 84B trained on the full Multi Med Bench mixture. As seen in Table 6, we observed that the model trained jointly on both report
为验证跨模态多任务联合训练带来的正向任务迁移效果,我们进行了消融实验:通过从任务混合集中排除MIMIC-CXR分类任务来训练Med-PaLM M 84B变体,并与完整Multi Med Bench混合集训练的Med-PaLM M 84B进行对比。如表6所示,我们观察到联合训练报告
Med-PaLM M 84B
Med-PaLM M 84B
No. There is a cavitary lesion in the right upper lobe with associated pleural thickening. The left lung appears clear. The car dio media st in al silhouette is unremarkable.
不。右上肺有一个空洞性病变,伴有胸膜增厚。左肺清晰。心脏纵隔轮廓未见异常。
Correctness: There is a cavitary lesion in the right upper lobe with associated pleural thickening, which is the most important of multiple important TB-related findings on this image.
正确性:右上肺存在一个空洞性病变伴胸膜增厚,这是该影像中最重要的多项结核相关表现之一。
Incorrectness: The left lung is not clear, with a small cavitary lesion in the left upper lobe related to TB. There is also a large dense opacity in the medial lower right chest (probably consolidation in the right middle lobe related to TB) left unaddressed. This opacity partially obscures the right cardiac silhouette.
左肺显示不清,左上叶可见与结核相关的小空洞病变。右胸内侧下部还存在一处未处理的大片致密影(可能为右中叶与结核相关的实变),该致密影部分遮蔽了右心缘轮廓。
Med-PaLM M 562B
Med-PaLM M 562B
No. There is cavitary lesion in the right upper lobe. There is also right hilar and media st in al adenopathy. There is scarring in the right middle lobe. There is no pleural effusion.
右上肺叶存在空洞性病变。右肺门及纵隔淋巴结肿大。右肺中叶可见瘢痕形成。未见胸腔积液。
Correctness: There is a cavitary lesion in the right upper lobe, which is the most important of multiple important TB-related findings on this image. There is no pleural effusion.
正确性:右上肺存在一个空洞性病变,这是该影像中多个重要结核相关表现中最关键的一处。未见胸腔积液。
Figure 3 | Evidence of emergent zero-shot multimodal medical reasoning with Med-PaLM M. Large Med-PaLM M models exhibit zero-shot CoT reasoning capability in identifying and describing tuberculosis related findings in chest X-ray images. The model is prompted with task-specific instructions and a text-only exemplar (without the corresponding image) to generate a report describing findings in the given X-ray image. Model predictions from Med-PaLM M 84B and 562B are shown together with the annotations from an expert radiologist. Both models correctly localized the major TB related cavitory lesion in the right upper lobe. However, both models did not address the small cavitory lesion in left upper lobe (Med-PaLM M 562B was considered better than Med-PaLM M 64B in this example as it also alluded to the opacity in the right middle lobe and did not make the incorrect statement of left lung being clear). Notably, Med-PaLM M 12B failed to generate a coherent report, indicating the importance of scaling for zero-shot COT reasoning.
图 3 | Med-PaLM M 展现的零样本多模态医学推理能力。大型 Med-PaLM M 模型在识别和描述胸部 X 光片中肺结核相关表现时,展现出零样本思维链 (CoT) 推理能力。模型通过任务指令和纯文本示例(不含对应图像)生成描述给定 X 光片表现的报告。Med-PaLM M 84B 和 562B 的预测结果与放射科专家的标注共同展示。两个模型均准确定位了右肺上叶的主要结核性空洞病变,但都未提及左肺上叶的小空洞病变(此例中 Med-PaLM M 562B 表现优于 64B 版本,因其同时提示了右肺中叶的混浊表现,且未错误声明左肺无异常)。值得注意的是,Med-PaLM M 12B 未能生成连贯报告,这表明模型规模对零样本思维链推理至关重要。
Partial correctness: Scarring in the right middle lobe, and right hilar and inferior media st in al adenopathy may both allude to the large dense opacity in the medial lower right chest (probably consolidation in the right middle lobe related to TB).
部分正确:右肺中叶瘢痕形成,以及右肺门和下纵隔淋巴结肿大,均可能指向右下胸部内侧的大片致密影(可能为与结核相关的右肺中叶实变)。
Incorrectness: The small cavitary in the left upper lobe lesion related to TB is unaddressed.
左上肺叶结核相关的小空洞病变未得到处理。
generation and classification has higher performance across the board on all report generation metrics. We also observe that the model trained only on chest X-ray report generation can generalize to abnormality detection in a zero-shot fashion with compelling performance, as evidenced by a higher macro-F1 score. This is another example of generalization to a novel task setting where the model learns to differentiate between types of abnormalities from training on the more complex report generation task.
在报告生成的各项指标上,生成与分类联合训练的模型均展现出更优性能。我们还观察到,仅针对胸部X光报告生成训练的模型能以零样本方式泛化至异常检测任务,并表现出色(宏观F1分数更高即为明证)。这再次体现了模型从更复杂的报告生成任务中学习后,能够泛化至新型任务场景(如区分各类异常情况)的能力。
6.3 Med-PaLM M performs encouragingly on radiology report generation across model scales
6.3 Med-PaLM M 在不同模型规模下均展现出令人鼓舞的放射学报告生成性能
To further understand the clinical applicability of Med-PaLM M, we conducted radiologist evaluations of model-generated chest X-ray reports (and reference human baselines). Under this evaluation framework, we observe encouraging quality of Med-PaLM M generated reports across model scales as detailed below.
为进一步评估Med-PaLM M的临床适用性,我们组织放射科医师对模型生成的胸部X光报告(及人类参考基线)进行评价。该评估框架显示,Med-PaLM M生成的报告质量随模型规模提升呈现积极趋势,具体如下。
6.3.1 Side-by-side evaluation
6.3.1 并行评估
In a side-by-side evaluation, four clinician raters ranked the quality of four radiology reports, comparing the radiologist-provided reference report from the MIMIC-CXR dataset with reports generated by different Med-PaLM M model scales (12B, 84B, and 562B).
在一次并行评估中,四位临床医生评审员对四份放射学报告的质量进行了排序,将MIMIC-CXR数据集中放射科医生提供的参考报告与不同规模的Med-PaLM M模型(12B、84B和562B)生成的报告进行了比较。
Figure 4a summarizes how often each rater ranked a report generated by one of the three Med-PaLM M variants or the reference report as the best among four candidate reports. Averaged over all four raters, the radiologist-provided reference report was ranked best in $37.14%$ of cases, followed by Med-PaLM M (84B)
图 4a 总结了每位评分者将三个 Med-PaLM M 变体生成的报告或参考报告评为四个候选报告中最佳报告的频率。四位评分者的平均结果显示,放射科医生提供的参考报告在 $37.14%$ 的案例中被评为最佳,其次是 Med-PaLM M (84B)。
Table 5 | Zero-shot generalization to two-view chest X-ray report generation. Med-PaLM M performance remains competitive on a novel two-view report generation task setup despite having not been trained with two visual inputs before. Med-PaLM M achieves SOTA results on clinical efficacy metrics for the two view report generation task.
| Metric | SOTA | Med-PaLM M (12B) | Med-PaLM M (84B) | Med-PaLM M (562B) |
| Micro-F1-14 | 44.20% | 49.80% | 50.54% | 48.85% |
| Macro-F1-14 | 30.70% | 37.69% | 37.78% | 37.29% |
| Micro-F1-5 | 56.70% | 54.49% | 56.37% | 54.36% |
| Macro-F1-5 | N/A | 48.33% | 51.23% | 48.49% |
| F1-RadGraph | 24.40% | 26.73% | 28.30% | 27.28% |
| BLEU-1 | 39.48% | 33.31% | 34.58% | 33.83% |
| BLEU-4 | 13.30% | 11.51% | 12.44% | 12.47% |
| ROUGE-L | 29.60% | 27.84% | 28.71% | 28.49% |
| CIDEr-D | 49.50% | 27.58% | 29.80% | 29.80% |
表 5 | 零样本泛化到双视角胸部X光报告生成。尽管此前未接受过双视觉输入训练,Med-PaLM M在新颖的双视角报告生成任务设置中仍保持竞争力,并在该任务的临床效能指标上达到SOTA水平。
| 指标 | SOTA | Med-PaLM M (12B) | Med-PaLM M (84B) | Med-PaLM M (562B) |
|---|---|---|---|---|
| Micro-F1-14 | 44.20% | 49.80% | 50.54% | 48.85% |
| Macro-F1-14 | 30.70% | 37.69% | 37.78% | 37.29% |
| Micro-F1-5 | 56.70% | 54.49% | 56.37% | 54.36% |
| Macro-F1-5 | N/A | 48.33% | 51.23% | 48.49% |
| F1-RadGraph | 24.40% | 26.73% | 28.30% | 27.28% |
| BLEU-1 | 39.48% | 33.31% | 34.58% | 33.83% |
| BLEU-4 | 13.30% | 11.51% | 12.44% | 12.47% |
| ROUGE-L | 29.60% | 27.84% | 28.71% | 28.49% |
| CIDEr-D | 49.50% | 27.58% | 29.80% | 29.80% |
Table 6 | Positive task transfer between CXR report generation and abnormality classification. We observe positive transfer as a result of multi-task training with Med-PaLM M model trained jointly on both chest X-ray report generation and classification tasks. It exhibits higher performance on report generation metrics compared to a Med-PaLM M model trained without chest X-ray report classification. We also observe that training on the chest X-ray report generation task alone enables Med-PaLM M to generalize to abnormality detection in a zero-shot fashion.
| Dataset | Metric | Med-PaLM M (84B) | Med-PaLM M (84B) No CXR classification |
| MIMIC-CXR | Micro-F1-14 | 53.56% | 52.94% |
| Macro-F1-14 | 39.83% | 38.92% | |
| Micro-F1-5 | 57.88% | 57.58% | |
| Macro-F1-5 | 51.60% | 51.32% | |
| F1-RadGraph | 26.71% | 26.08% | |
| BLEU-1 | 32.31% | 31.72% | |
| BLEU-4 | 11.31% | 10.87% | |
| ROUGE-L | 27.29% | 26.67% | |
| CIDEr-D | 26.17% | 25.17% | |
| MIMIC-CXR (5 conditions) | Macro-AUC | 78.35% | 73.88% |
| Macro-F1 | 36.83% | 43.97% |
表 6 | 胸部X光报告生成与异常分类任务间的正向迁移。我们观察到,通过Med-PaLM M模型在胸部X光报告生成和分类任务上的多任务联合训练,产生了正向迁移效应。与未进行胸部X光报告分类训练的Med-PaLM M模型相比,该模型在报告生成指标上表现出更高性能。我们还发现,仅通过胸部X光报告生成任务的训练,就能使Med-PaLM M以零样本方式泛化至异常检测任务。
| 数据集 | 指标 | Med-PaLM M (84B) | Med-PaLM M (84B) 无CXR分类 |
|---|---|---|---|
| MIMIC-CXR | Micro-F1-14 | 53.56% | 52.94% |
| Macro-F1-14 | 39.83% | 38.92% | |
| Micro-F1-5 | 57.88% | 57.58% | |
| Macro-F1-5 | 51.60% | 51.32% | |
| F1-RadGraph | 26.71% | 26.08% | |
| BLEU-1 | 32.31% | 31.72% | |
| BLEU-4 | 11.31% | 10.87% | |
| ROUGE-L | 27.29% | 26.67% | |
| CIDEr-D | 26.17% | 25.17% | |
| MIMIC-CXR (5种病症) | Macro-AUC | 78.35% | 73.88% |
| Macro-F1 | 36.83% | 43.97% |
which was ranked best in $25.78%$ of cases, and the other two model scales, 12B and 562B, which were ranked best in $19.49%$ and $17.59%$ of cases respectively.
在25.78%的情况下排名最佳,而其他两种模型规模(12B和562B)分别以19.49%和17.59%的比例位列最优。
To enable a direct comparison of reports generated by each Med-PaLM M model scale to the radiologistprovided reference report, we derived pairwise preferences from the four-way ranking and provided a breakdown for each rater and model scale in Figure 4b. Averaged over all four raters, Med-PaLM M 84B was preferred over the reference report in $40.50%$ of cases, followed by the other two model scales, 12B and 562B, which were preferred over the reference report in $34.05%$ and $32.00%$ of cases, respectively.
为了直接比较每个Med-PaLM M模型规模生成的报告与放射科医生提供的参考报告,我们从四向排名中得出成对偏好,并在图4b中为每位评分者和模型规模提供了细分数据。四位评分者的平均结果显示,Med-PaLM M 84B在$40.50%$的案例中优于参考报告,其次是另外两个模型规模12B和562B,它们分别以$34.05%$和$32.00%$的案例表现优于参考报告。
6.3.2 Independent evaluation
6.3.2 独立评估
We report the rates of omissions and errors radiologists identified in findings paragraphs generated by MedPaLM M. Figure 5 provides breakdowns by model scales (12B, 84B, 562B). We observed different trends for omissions and errors. For omissions, we observed the lowest rate of 0.12 (95% CI, 0.10 - 0.15) omissions per report on average for both the Med-PaLM M 12B and 84B models, followed by 0.13 (95% CI, 0.11 - 0.16) for the 562B model.
我们报告了放射科医师在MedPaLM M生成的检查结果段落中发现的遗漏和错误率。图5展示了不同模型规模(12B、84B、562B)的细分数据。我们观察到遗漏和错误呈现出不同的趋势。在遗漏方面,Med-PaLM M 12B和84B模型的表现最佳,平均每份报告遗漏0.12次(95% CI, 0.10 - 0.15),而562B模型为0.13次(95% CI, 0.11 - 0.16)。
In contrast, we measured the lowest mean error rate of 0.25 (95% CI, 0.22 - 0.28) for Med-PaLM M 84B, followed by 0.28 (95% CI, 0.24 - 0.31) for Med-PaLM M 12B and 0.29 ( $95%$ CI, 0.25 - 0.32) for the 562B model. Notably, this error rate is comparable to those reported for human radiologists baselines on the MIMIC-CXR
相比之下,我们测得 Med-PaLM M 84B 的平均错误率最低,为 0.25 (95% CI, 0.22 - 0.28),其次是 Med-PaLM M 12B 的 0.28 (95% CI, 0.24 - 0.31) 和 562B 模型的 0.29 (95% CI, 0.25 - 0.32)。值得注意的是,这一错误率与 MIMIC-CXR 上报告的放射科医生基线水平相当。

Figure 4 | Side-by-side human evaluation. Four clinician raters ranked the quality of four radiology reports in a side-by-side evaluation, comparing the radiologist-provided reference report from MIMIC-CXR with reports generated by different Med-PaLM M model scale variants (12B, 84B, 562B).
图 4 | 并行人工评估。四位临床医生评审员在并行评估中对四份放射学报告的质量进行排名,比较了MIMIC-CXR提供的放射科医生参考报告与不同规模Med-PaLM M模型变体(12B、84B、562B)生成的报告。
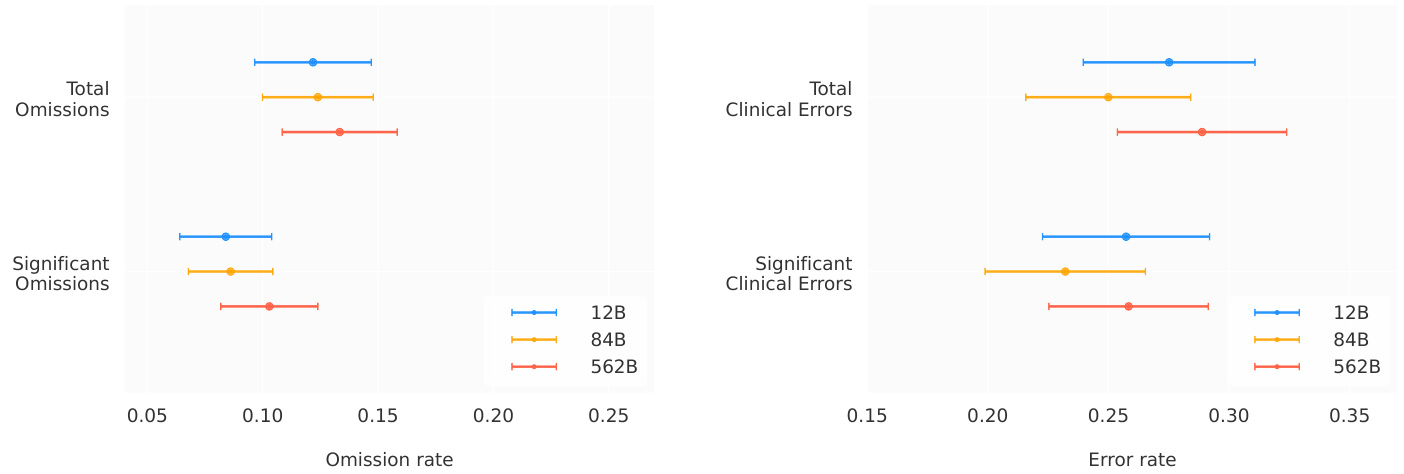
Figure 5 | Independent human evaluation. Rates of omissions and clinical errors identified by clinician raters in radiology reports generated by Med-PaLM M. Clinical errors are those related to the presence, location or severity of a clinical finding.
图 5 | 独立人类评估。临床评审人员在 Med-PaLM M 生成的放射学报告中发现的遗漏率和临床错误率。临床错误指与临床发现的存现、位置或严重程度相关的错误。
dataset in a prior study [14].
先前研究中的数据集[14]。
It is important to mention that our analysis is limited to errors of clinical relevance, ensuring a specific focus on clinical interpretation. This includes those errors related to the presence, location or severity of a clinical finding. Example of non-clinical errors are passages referring to views or prior studies not present, which stem from training artifacts.
需要说明的是,我们的分析仅限于具有临床意义的错误,确保聚焦于临床解读。这包括与临床表现的存在、位置或严重程度相关的错误。非临床错误的例子包括提及不存在的视图或既往研究,这些源自训练过程中的伪影。
These trends across model scales were identical for the subset of omissions and errors that were marked as significant by radiologist raters. We refer the reader to Table A.8 for an overview of error and omission rates, including non-clinical errors.
这些趋势在不同模型规模上对于放射科医生评定为显著的遗漏和错误子集同样适用。有关错误和遗漏率(包括非临床错误)的概述,请参阅表 A.8。
In Figure 6, we illustrate a qualitative example of chest X-ray reports generated by Med-PaLM M across three model sizes along with the target reference report. For this example, our panel of radiologists judged the Med-PaLM M 12B report to have two clinically significant errors and one omission, the Med-PaLM M 84B report to have zero errors and zero omissions, and the Med-PaLM M 562B report to have one clinically insignificant errors and no omissions.
在图 6 中,我们展示了由 Med-PaLM M 生成的胸部 X 光报告与目标参考报告的定性比较示例,涵盖三种模型规模。对于此案例,我们的放射科医师小组判定:Med-PaLM M 12B 版本存在两处临床显著错误和一处遗漏,Med-PaLM M 84B 版本零错误零遗漏,Med-PaLM M 562B 版本存在一处临床非显著错误且无遗漏。
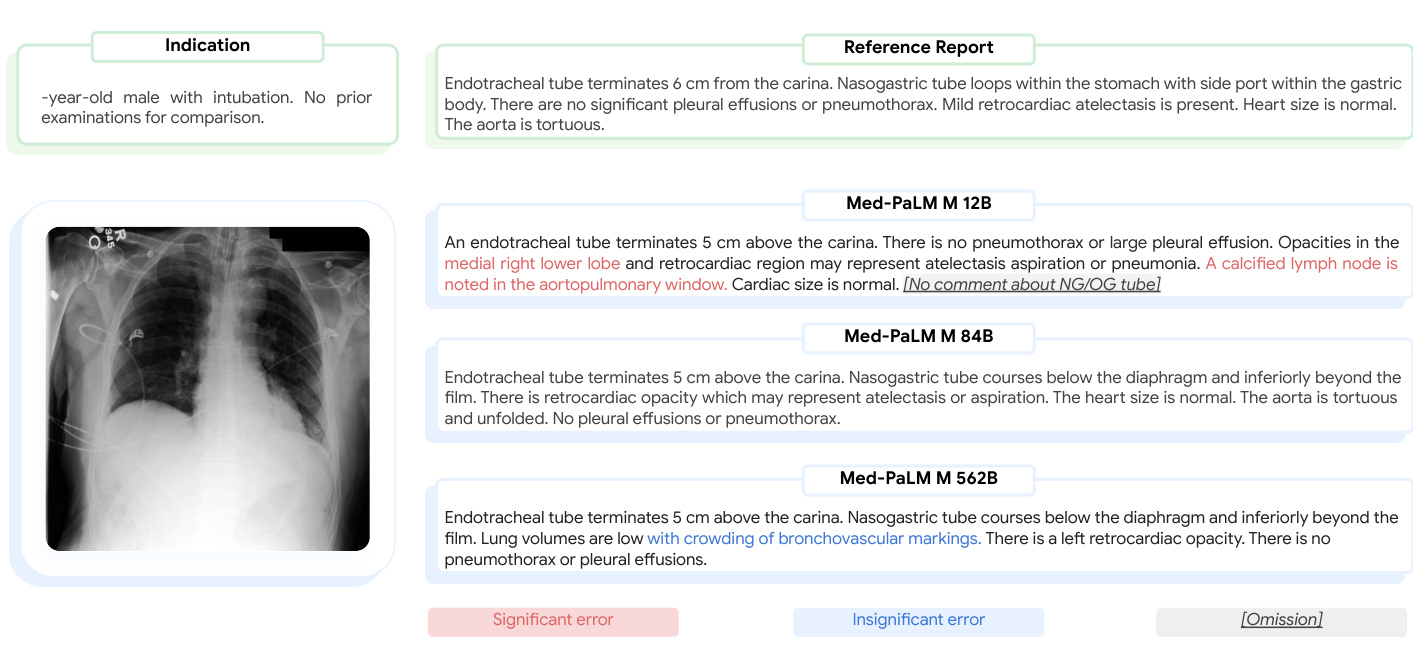
Figure 6 | Qualitative example of reference and Med-PaLM M generated chest $\mathbf{x}$ -ray reports. We present a qualitative example of chest X-ray reports generated by Med-PaLM M across model scales along with the target reference report. In this example, a panel of radiologists adjudicated the Med-PaLM M 12B report to have two clinically significant errors and one omission, the Med-PaLM M 84B report to have zero errors and zero omissions, and the Med-PaLM M 562B report to have one clinically insignificant error and no omissions.
图 6 | 参考报告与 Med-PaLM M 生成的胸部 $\mathbf{x}$ 光报告定性对比示例。我们展示了不同规模 Med-PaLM M 模型生成的胸部 X 光报告与目标参考报告的定性对比案例。本例中,放射科医师小组判定:Med-PaLM M 12B 报告存在两处临床显著错误和一处遗漏,Med-PaLM M 84B 报告零错误零遗漏,Med-PaLM M 562B 报告存在一处临床非显著错误且无遗漏。
7 Discussion
7 讨论
To the best of our knowledge, Med-PaLM M is the first demonstration of a generalist biomedical AI system that can interpret a wide range of medical modalities, perform competently (including near or exceeding prior SOTA) on a diverse array of tasks, and generalize to unseen biomedical concepts and tasks. This potentially opens up new possibilities in applications spanning scientific discovery to care delivery. We elaborate on the implications of this development as well as the challenges and limitations below.
据我们所知,Med-PaLM M 是首个通用生物医学AI系统的示范案例,能够解读多种医学模态数据,在多样化任务中表现优异(接近或超越先前SOTA水平),并能泛化至未见过的生物医学概念和任务。这为从科学发现到医疗护理的应用领域开辟了新的可能性。下文我们将详细探讨这一进展的意义、面临的挑战与局限性。
Lack of benchmarks a key bottleneck for the development of generalist biomedical AI AI progress to date has largely been catalyzed by the development of high quality benchmarks. While there exists several single-task biomedical AI datasets, there have been limited attempts to unify them and create benchmarks for the development of generalist biomedical AI systems. Our curation of Multi Med Bench is a step towards addressing this unmet need. However, the benchmark has several important limitations including limited size of the individual datasets (a cumulative size of $\tilde{1}$ million samples) and limited modality and task diversity (e.g., lacking life sciences such as transcript om ics and proteomics). Another key barrier to developing models for use across an even wider variety of biomedical data types is the lack of large scale multimodal datasets, which would permit joint learning and alignment of the modality-specific encoders with the decoder.
缺乏基准测试是开发生物医学通用AI的关键瓶颈
迄今为止,AI的进步很大程度上得益于高质量基准测试的发展。虽然已有多个单任务生物医学AI数据集,但将它们统一起来并为开发生物医学通用AI系统创建基准测试的尝试仍然有限。我们整理的Multi Med Bench是满足这一未满足需求的一步。然而,该基准测试存在几个重要限制,包括单个数据集的规模有限(累计约$\tilde{1}$百万样本)以及模态和任务多样性不足(例如缺少转录组学和蛋白质组学等生命科学领域)。另一个阻碍开发适用于更广泛生物医学数据类型的模型的关键障碍是缺乏大规模多模态数据集,这些数据集可以实现模态特定编码器与解码器的联合学习和对齐。
Importance of medical finetuning and specialization PaLM-E is a highly capable generalist AI model as evidenced by its SOTA performance on a wide range of vision-language and embodied robotics tasks. Yet, its out-of-the-box performance on Multi Med Bench was poor and Med-PaLM M outperforms it by a wide margin across model scales. This result suggests that finetuning with domain-specific biomedical data is critical to achieving good performance on biomedical tasks, perhaps due to the distribution shift presented by the domain overall compared to the plethora of non-medical tasks and modalities.
医疗领域微调与专业化的重要性
PaLM-E 作为一款全能型AI模型,在各类视觉语言和具身机器人任务中展现的SOTA性能已充分证明其卓越能力。然而该模型在Multi Med Bench基准测试中的开箱性能表现不佳,且Med-PaLM M在不同模型规模下均大幅领先。这一结果表明:针对生物医学领域数据进行的专项微调对获得优异表现至关重要,其根本原因可能在于医学领域整体与非医疗任务及模态之间存在显著的数据分布偏移。
Scaling multimodal AI models is challenging In the language domain, scaling the model has led to leapfrog improvements in performance and emergent capabilities. However, our preliminary experiments suggest this is likely more challenging for multimodal generalist models in the biomedical task domain due to the medical data scarcity. Given the wide array of modalities and tasks such generalist models are expected to understand and tackle, it is crucial that the encoders for such diverse modalities are scaled jointly with the language model. Otherwise, for tasks that require interpretation of data from a combination of modalities, the performance will end up being bottle necked by the weakest encoder. We see evidence of this in medical image classification tasks such as mammography and dermatology where scaling the language model component has little effect on the task performance as the potential key bottleneck is the vision encoder. It is possible that the small volume of medical data in Multi Med Bench is not be sufficient to effectively adapt a ViT pretrained on natural images to the medical domain, thereby limiting the benefits of model scaling. As such, our study only provides some initial insights on the effect of model scaling on biomedical task performance. Future research is needed to fully understand the effect of model scaling by teasing apart the scaling effect of the language model from that of modality-specific encoders, with sufficient amounts of biomedical data.
扩展多模态AI模型具有挑战性
在语言领域,扩展模型带来了性能和涌现能力的飞跃式提升。然而,我们的初步实验表明,由于医学数据稀缺,这对于生物医学任务领域的多模态通用模型可能更具挑战性。鉴于这类通用模型需要理解和处理多种模态与任务的广泛组合,关键是要让这些多样化模态的编码器与大语言模型协同扩展。否则,对于需要解释多模态组合数据的任务,性能最终会被最弱的编码器所限制。我们在乳腺X光摄影和皮肤病学等医学图像分类任务中观察到了这一现象——扩展语言模型组件对任务性能几乎无影响,因为潜在的关键瓶颈在于视觉编码器。Multi Med Bench中少量的医学数据可能不足以有效将基于自然图像预训练的ViT适配到医学领域,从而限制了模型扩展的收益。因此,我们的研究仅提供了关于模型扩展对生物医学任务性能影响的初步见解。未来需要进一步研究,通过分离语言模型与特定模态编码器的扩展效应,并基于足量生物医学数据,以全面理解模型扩展的影响。
Technical considerations for generalist biomedical AI Med-PaLM M builds on state-of-the-art vision and language components such as ViT and PaLM. Yet, putting them together requires careful considerations around token lengths allocated to visual encoder outputs, total context length of the model, sampling strategies, training data mixtures and so forth. Further, simple, yet important techniques such as the use of one-shot training with dummy image tokens make an important difference in the quality and compute efficiency of the final model. With increasing generality of the AI system, the number of details requiring careful consideration tends to increase as well. We also note that Med-PaLM M architecture as setup currently is not optimal for few-shot in-context learning.
通用生物医学AI的技术考量
Med-PaLM M基于最先进的视觉与语言组件(如ViT和PaLM)构建,但整合这些组件需谨慎考虑视觉编码器输出的token分配长度、模型总上下文长度、采样策略、训练数据混合比例等因素。此外,诸如使用带虚拟图像token的单样本训练等简单却重要的技术,对最终模型的质量与计算效率有显著影响。随着AI系统通用性的提升,需细致考量的技术细节往往同步增加。我们同时指出,当前Med-PaLM M架构在少样本上下文学习方面尚未达到最优状态。
Progress in AI for radiology report generation Our evaluation by radiologists of Med-PaLM M generated radiology reports suggests encouraging performance of the model on a challenging multimodal task. In up to $40.50%$ of the cases, a Med-PaLM M generated report was preferred over the human-generated reference report. Further, the average number of clinically significant errors within the model responses is comparable to those reported for human-generated reports in prior studies [14] on the same dataset. These promising results underpin rapid development in the task of automatic radiology report generation and suggest the potential for clinical utility in the future.
放射学报告生成领域的AI进展
放射科医师对Med-PaLM M生成的放射学报告评估表明,该模型在具有挑战性的多模态任务中表现令人鼓舞。在高达$40.50%$的案例中,Med-PaLM M生成的报告比人工撰写的参考报告更受青睐。此外,模型响应中临床显著错误的平均数量与先前同一数据集研究中[14]报告的人工撰写错误率相当。这些积极结果支撑了自动放射学报告生成任务的快速发展,并预示着未来潜在的临床应用价值。
Generalist agents are not the only approach to multimodal biomedical AI While generalist biomedical AI systems offer exciting possibilities [43], there are other approaches to developing multimodal biomedical AI systems that might be more applicable depending on data availability, pretrained models, compute and application scenarios. These include leveraging frozen encoders with adapter layers [75] to glue together a multimodal biomedical AI system or developing LLMs that can interface with specialist biomedical encoders or task-specific agents through tool use [76].
通用智能体并非多模态生物医学AI的唯一途径
尽管通用生物医学AI系统展现出令人兴奋的可能性 [43],但根据数据可用性、预训练模型、算力和应用场景的不同,开发多模态生物医学AI系统还存在其他可能更适用的方法。这些方法包括:利用带有适配器层 (adapter layers) 的冻结编码器 (frozen encoders) [75] 来构建多模态生物医学AI系统,或是开发能够通过工具调用 [76] 与专业生物医学编码器或任务特定智能体交互的大语言模型。
Considerations for real-world applications of generalist biomedical AI While the development of generally capable biomedical AI systems is exciting, for such systems to be useful in practice or opening the door to new applications, they need to match or exceed specialized, single-task models or otherwise reach clinically applicable levels of performance. While beyond the scope of this work, the progress here necessitates careful considerations of safety and equity in the development and validation of such systems.
通用生物医学AI在实际应用中的考量
虽然通用生物医学AI系统的开发令人振奋,但要让这些系统在实践中发挥作用或开启新的应用领域,它们需要达到或超越专门化的单任务模型,或者至少达到临床适用的性能水平。尽管这超出了本研究的范围,但当前的进展要求我们在开发和验证此类系统时,必须审慎考虑安全性与公平性。
8 Perspective on Generalist Biomedical AI
8 通用生物医学AI的展望
Reaching near or above SOTA on a diverse range of biomedical tasks with a single set of model weights is a noteworthy milestone for the development of generalist biomedical AI systems. While human clinicians can train for “general practice” [77], helpful sub specialty-specific expertise is often found in different experts [78], to whom non-specialist clinicians may refer for specialist opinions in the course of care. It is also commonplace for multiple physician specialities to work together in care delivery. We envisage a similar future for biomedical AI where generalist and specialist AI systems interact and collaborate together with expert clinicians and researchers in a tight feedback loop to tackle grand challenges in bio medicine.
在多种生物医学任务上达到接近或超越当前最优水平 (SOTA) ,且仅使用单一模型权重集,这是通用生物医学AI系统发展的重要里程碑。虽然人类临床医生可以接受"全科医疗"培训 [77] ,但实用的亚专科专业知识通常分散在不同专家身上 [78] ,非专科临床医生在诊疗过程中会向这些专家寻求专科意见。多个医师专科协同开展诊疗工作也是普遍做法。我们预见生物医学AI将出现类似的发展图景:通用AI系统与专科AI系统将形成紧密反馈循环,与临床专家和研究人员互动协作,共同应对生物医学领域的重大挑战。
Our finding, of a single generalist biomedical AI that reaches compelling performance across disparate tasks and contexts, hints at new frontiers for impact in applications. This includes the potential for near zero-shot insight in new domains, as a tool for discovery integrating insights from distinct areas of bio medicine, and as a common point of assistance providing access to expertise from many different fields.
我们的研究发现,一个单一通用的生物医学AI能够在不同任务和情境中达到引人注目的性能,这暗示了其在应用领域的新前沿。包括在新领域中近乎零样本的洞察潜力,作为整合生物医学不同领域见解的发现工具,以及作为提供多领域专业知识的共同辅助点。
9 Conclusion
9 结论
Medicine is a multidisciplinary endeavour. Generalist biomedical AI systems that effectively assimilate and encode multimodal medical data at scale and rapidly adapt to new clinical contexts are likely to be the foundation of next generation learning health systems and make healthcare more accessible, efficient, equitable and humane. While further development and rigorous validation is needed, we believe Med-PaLM M represents an important step towards the development of such generalist biomedical AI.
医学是一门多学科交叉的学科。能够有效整合并编码大规模多模态医疗数据、快速适应新临床场景的通才型生物医学AI系统,很可能成为下一代学习型医疗体系的基石,使医疗保健服务更具可及性、高效性、公平性和人文关怀。尽管仍需进一步开发和严格验证,我们认为Med-PaLM M代表了通向这类通才型生物医学AI发展的重要一步。
Acknowledgments
致谢
This project was an extensive collaboration between many teams at Google Research and Google DeepMind. We thank Andrew Sellergren, Yuan Liu, Michael Howell, Julie Wang, Sho Kannan, Christine Kingsley, Roy Lee, Naama Hammel, Jay Hartford, Preeti Singh, Kavita Kulkarni, Gavriel Goidel, Anil Palepu, Si Wai Man, Amy Wang, Sami Lachgar, Lauren Winer, Maggie Shiels, Annisah Um’rani, John Guilyard, Shravya Shetty and Evan Rapoport for their valuable insights and feedback during our research. We are also grateful to Karen DeSalvo, Zoubin Ghahramani, James Manyika, and Jeff Dean for their support during the course of this project.
本项目是Google Research与Google DeepMind多个团队之间的广泛合作成果。我们感谢Andrew Sellergren、Yuan Liu、Michael Howell、Julie Wang、Sho Kannan、Christine Kingsley、Roy Lee、Naama Hammel、Jay Hartford、Preeti Singh、Kavita Kulkarni、Gavriel Goidel、Anil Palepu、Si Wai Man、Amy Wang、Sami Lachgar、Lauren Winer、Maggie Shiels、Annisah Um'rani、John Guilyard、Shravya Shetty和Evan Rapoport在研究过程中提供的宝贵见解与反馈。同时感谢Karen DeSalvo、Zoubin Ghahramani、James Manyika和Jeff Dean对本项目的全程支持。
Data Availability
数据可用性
The benchmark used for training and evaluation in this study, Multi Med Bench, comprises de-identified datasets that are all open source. We present an overview of datasets in Table 1.
本研究用于训练和评估的基准Multi Med Bench包含全部开源且经过去标识化的数据集。各数据集概览如 表1: 所示。
Code Availability
代码可用性
We will not be able to open source the large languag