BIOMEDGPT: OPEN MULTIMODAL GENERATIVE PRE-TRAINED TRANSFORMER FOR BIO MEDICINE
BIOMEDGPT: 面向生物医学领域的开源多模态生成式预训练Transformer
Yizhen $\mathbf{L}\mathbf{u0}^{1* }$ , Jiahuan Zhang1∗, Siqi $\mathbf{Fan}^{1* }$ , Kai $\mathbf{Yang^{1* }}$ , Yushuai $\mathbf{W}\mathbf{u}^{1* }$ , $\mathbf{M}\mathbf{u}\mathbf{Q}\mathbf{i}\mathbf{a}\mathbf{o}^{2\dagger}$ , Zaiqing Nie1,2† Institute for AI Industry Research (AIR), Tsinghua University 1 PharMolix Inc.2 yz-luo22@mails.tsinghua.edu.cn, mqiao@pharmolix.com {zhang jia huan, fansiqi, yangkai, zaiqing}@air.tsinghua.edu.cn
Yizhen $\mathbf{L}\mathbf{u0}^{1* }$, Jiahuan Zhang1∗, Siqi $\mathbf{Fan}^{1* }$, Kai $\mathbf{Yang^{1* }}$, Yushuai $\mathbf{W}\mathbf{u}^{1* }$, $\mathbf{M}\mathbf{u}\mathbf{Q}\mathbf{i}\mathbf{a}\mathbf{o}^{2\dagger}$, Zaiqing Nie1,2†
清华大学智能产业研究院(AIR)1
PharMolix Inc.2
yz-luo22@mails.tsinghua.edu.cn, mqiao@pharmolix.com
{zhang jia huan, fansiqi, yangkai, zaiqing}@air.tsinghua.edu.cn
ABSTRACT
摘要
Foundation models (FMs) have exhibited remarkable performance across a wide range of downstream tasks in many domains. Nevertheless, general-purpose FMs often face challenges when confronted with domain-specific problems, due to their limited access to the proprietary training data in a particular domain. In bio medicine, there are various biological modalities, such as molecules, proteins, and cells, which are encoded by the language of life and exhibit significant modality gaps with human natural language. In this paper, we introduce BioMedGPT, an open multimodal generative pre-trained transformer (GPT) for bio medicine, to bridge the gap between the language of life and human natural language. BioMedGPT allows users to easily “communicate” with diverse biological modalities through free text, which is the first of its kind. BioMedGPT aligns different biological modalities with natural language via a large generative language model, namely, BioMedGPT-LM. We publish BioMedGPT-10B, which unifies the feature spaces of molecules, proteins, and natural language via encoding and alignment. Through fine-tuning, BioMedGPT-10B outperforms or is on par with human and significantly larger general-purpose foundation models on the biomedical QA task. It also demonstrates promising performance in the molecule QA and protein QA tasks, which could greatly accelerate the discovery of new drugs and therapeutic targets. In addition, BioMedGPTLM-7B is the first large generative language model based on Llama2 in the biomedical domain, therefore is commercial friendly. Both BioMedGPT-10B and BioMedGPT-LM-7B are open-sourced to the research community. In addition, we publish the datasets that are meticulously curated for the alignment of multi-modalities, i.e., PubChemQA and UniProtQA. All the models, codes, and datasets are available at https://github.com/PharMolix/OpenBioMed.
基础模型 (FMs) 在多个领域的广泛下游任务中展现出卓越性能。然而,通用基础模型在面临特定领域问题时往往存在局限性,这源于其对专有训练数据的获取受限。在生物医学领域,存在分子、蛋白质、细胞等多种生物模态,它们由生命语言编码,与人类自然语言存在显著模态差异。本文提出BioMedGPT——一个面向生物医学的开源多模态生成式预训练Transformer (GPT),旨在弥合生命语言与人类自然语言之间的鸿沟。BioMedGPT首次实现通过自由文本与多种生物模态"对话"的能力。该系统通过大语言模型BioMedGPT-LM,将不同生物模态与自然语言对齐。我们发布了BioMedGPT-10B,其通过编码和对齐技术统一了分子、蛋白质与自然语言的特征空间。经微调后,BioMedGPT-10B在生物医学问答任务上表现优于或持平人类专家,且显著超越规模更大的通用基础模型。在分子问答和蛋白质问答任务中也展现出卓越性能,有望大幅加速新药与治疗靶点的发现。此外,BioMedGPT-LM-7B是生物医学领域首个基于Llama2架构的大规模生成式语言模型,具备商业化友好特性。BioMedGPT-10B与BioMedGPT-LM-7B均已向研究社区开源。我们还发布了专为多模态对齐精心构建的数据集PubChemQA和UniProtQA。所有模型、代码及数据集详见https://github.com/PharMolix/OpenBioMed。
Keywords Large Language Model $\cdot$ Bio medicine $\cdot$ Generation $\cdot$ Alignment $\cdot$ Multi-Modality
关键词 大语言模型 (Large Language Model) $\cdot$ 生物医学 $\cdot$ 生成 $\cdot$ 对齐 $\cdot$ 多模态
1 Introduction
1 引言
Foundation models (FMs) are large AI models trained on enormous amounts of unlabelled data through self-supervised learning. FMs such as ChatGPT [OpenAI, 2022], Bard [Google, 2023], and Chinchilla [Hoffmann et al., 2022] have showcased impressive general intelligence across a broad spectrum of tasks. Most recently, Meta introduced Llama2 [Touvron et al., 2023a], a family of pre-trained and instruction-tuned large language models (LLMs), demonstrating prevailing results over existing open-source LLMs and on-par performance with some of the closed-source models based on human evaluations in terms of helpfulness and safety.
基础模型 (Foundation Model, FM) 是通过自监督学习在大量无标注数据上训练的大型 AI 模型。诸如 ChatGPT [OpenAI, 2022]、Bard [Google, 2023] 和 Chinchilla [Hoffmann et al., 2022] 等基础模型已在广泛任务中展现出卓越的通用智能。最近,Meta 推出了 Llama2 [Touvron et al., 2023a] 系列预训练及指令微调的大语言模型,在人类评估的有用性和安全性方面,其表现优于现有开源大语言模型,并与部分闭源模型相当。
However, these general-purpose FMs are typically trained on internet-scale generic public datasets, and their depth of knowledge within particular domains is restricted by the lack of access to proprietary training data. To solve this challenge, domain-specific FMs are becoming more prevalent, and attracting tremendous attention to their widespread applications in many fields. For instance, Bloomberg GP T [Wu et al., 2023a] is a large-scale GPT model for finance.
然而,这些通用基础模型(FM)通常是在互联网规模的通用公共数据集上训练的,它们在特定领域的知识深度受到缺乏专有训练数据的限制。为解决这一挑战,领域专用基础模型正变得越来越普遍,并因其在许多领域的广泛应用而受到极大关注。例如,Bloomberg GPT [Wu et al., 2023a] 就是一款面向金融领域的大规模GPT模型。
ChatLaw [Cui et al., 2023] fine-tunes Llama on a legal database and shows promising results on downstream legal applications.
ChatLaw [Cui et al., 2023] 在法律数据库上对 Llama 进行微调,并在下游法律应用中展现出良好效果。
Recently, research works about FMs in the biomedical domain have emerged. BioMedLM, an open-source GPT model with 2.7 billion parameters, is trained exclusively using PubMed abstracts and the PubMed Central section from the Pile data [Gao et al., 2020]. PMC-Llama [Wu et al., 2023b], a biomedical FM fine-tuned from Llama-7B with millions of biomedical publications, has demonstrated superior understanding of biomedical knowledge over general-purpose FMs. Biomedical FMs excel in grasping human language but struggle with comprehending diverse biomedical modalities, including molecular structures, protein sequences, pathways, and cell transcript om ics. Bio medicine, akin to human and computers, has its own distinct “languages”, like the molecular language, which employs molecular grammars to generate molecules or polymers [Guo et al., 2023]. Recent research efforts have been devoted to harnessing large-scale pre-trained language models for learning these multi-modalities. To bridge the modality gap, Bio Translator [Xu et al., 2023] develops a multilingual translation framework to translate text descriptions to non-text biological data instances. In particular, Bio Translator fine-tunes PubMedBERT [Gu et al., 2020] on existing biomedical ontologies and utilizes the resulting domain-specific model to encode textual descriptions. Bio Translator bridges different forms of biological data by projecting encoded text data and non-text biological data into a shared embedding space via contrastive learning.
近期,生物医学领域关于基础模型(FM)的研究成果不断涌现。BioMedLM作为一款开源的27亿参数GPT模型,其训练数据仅采用Pile数据集中的PubMed摘要和PubMed Central文献[Gao et al., 2020]。基于Llama-7B框架、使用数百万篇生物医学文献微调而成的PMC-Llama[Wu et al., 2023b],在生物医学知识理解方面展现出超越通用基础模型的优势。虽然生物医学基础模型擅长处理人类语言,但在理解分子结构、蛋白质序列、通路和细胞转录组等多模态生物医学数据时仍存在局限。与人类和计算机类似,生物医学也有其独特的"语言体系",例如运用分子语法生成分子或聚合物的分子语言[Guo et al., 2023]。当前研究重点聚焦于利用大规模预训练语言模型学习这些多模态表征。为弥合模态鸿沟,Bio Translator[Xu et al., 2023]开发了多语言翻译框架,可将文本描述转化为非文本生物数据实例。该研究通过在现有生物医学本体上微调PubMedBERT[Gu et al., 2020]获得领域专用模型来编码文本描述,并借助对比学习将编码后的文本数据与非文本生物数据映射到共享嵌入空间,从而实现不同形式生物数据的关联。
The inherent laws of nature and the evolution of life, dominated by the organization and interaction of atoms and molecules, employ the language of life to constitute the first principle in bio medicine. Specifically, the language of life describes how information is encoded in logical ways to ensure proper functionalities underlying all of life. For example, the nucleic acids of DNA encode genetic information while the amino acid sequence contains codes to translate that information into protein structures and functions. In addition, proteins are the key building blocks in any organism, from single cells to much more complex organisms such as animals. On the other hand, humans have accumulated extensive biomedical knowledge over centuries, which are usually described in natural language and represented in the forms of knowledge graphs, text documents, and experimental results. There are still many territories yet to be explored by humans in order to understand the fundamental codes, i.e., the language of life. For example, as of June 2023, UniProtKB/TrEMBL [Consortium, 2022] records more than 248M proteins, but only $0.2%$ of them are well studied and manually annotated by human experts. Recently, specialized AI models like ESM-2 [Lin et al., 2022] have shown promise in mining the substantial uncharted regions in bio medicine. However, these black-box models are incapable of providing scientific insights with human interpret able language. More recently, GPT-4 [OpenAI, 2023] demonstrates great success in comprehending not only natural language but also more structured data, such as graphs, tables, and diagrams, illuminating an opportunity to bridge the gap between the language of life and natural language, and therefore revolutionize scientific research.
自然界的固有规律与生命演化,由原子和分子的组织与相互作用主导,运用生命语言构成了生物医学的第一性原理。具体而言,生命语言描述了信息如何以逻辑方式编码,以确保支撑所有生命体的基础功能。例如,DNA的核酸编码遗传信息,而氨基酸序列则包含将这些信息翻译为蛋白质结构与功能的密码。此外,蛋白质是从单细胞到动物等更复杂生物体的关键组成部分。另一方面,人类几个世纪以来积累了大量的生物医学知识,这些知识通常以自然语言描述,并以知识图谱、文本文档和实验结果等形式呈现。为了理解生命的基本密码(即生命语言),人类仍有许多未知领域有待探索。例如,截至2023年6月,UniProtKB/TrEMBL [Consortium, 2022] 记录了超过2.48亿种蛋白质,但其中仅有0.2%经过深入研究并由人类专家手动注释。最近,ESM-2 [Lin et al., 2022] 等专业AI模型在开发生物医学中大量未探索区域方面展现出潜力。然而,这些黑盒模型无法用人类可解释的语言提供科学见解。更近期的GPT-4 [OpenAI, 2023] 不仅在理解自然语言方面取得巨大成功,还能处理更结构化的数据(如图表、表格和示意图),这为弥合生命语言与自然语言之间的鸿沟提供了契机,从而可能彻底改变科学研究。
To achieve this over arching and ambitious goal, we develop BioMedGPT, a novel framework to bridge the language of life and human natural language using large-scale pre-trained language models. Figure 1 shows the overview of BioMedGPT. We build a biomedical language model, BioMedGPT-LM, by fine-tuning existing general-purpose LLMs with large-scale biomedical corpus. Built on top of BioMedGPT-LM, BioMedGPT is designed to unify the language of life, encoding biological building blocks such as molecules, proteins, and cells, as well as natural languages, describing human knowledge in the forms of knowledge graphs, texts, and experimental results. Alignment and translation between different biological modalities and human natural language are performed via neural networks and self-supervised learning techniques. Compared with prior work, BioMedGPT enjoys the benefits of larger GPT type of LLMs and a substantial amount of untapped texts in the biomedical research field. Within the unified feature space of different modalities, BioMedGPT can flexibly support free-text inquiries, digest multimodal inputs, and versatile ly power a wide range of downstream tasks without end-to-end fine-tuning.
为实现这一宏大而雄心勃勃的目标,我们开发了BioMedGPT——一个通过大规模预训练语言模型连接生命语言与人类自然语言的新型框架。图1展示了BioMedGPT的整体架构。我们通过使用大规模生物医学语料库微调现有通用大语言模型,构建了生物医学语言模型BioMedGPT-LM。基于BioMedGPT-LM构建的BioMedGPT旨在统一生命语言(编码分子、蛋白质、细胞等生物构件)与自然语言(以知识图谱、文本和实验结果形式描述人类知识)。通过神经网络和自监督学习技术,实现不同生物模态与人类自然语言之间的对齐与转换。与现有工作相比,BioMedGPT兼具更大规模的GPT类大语言模型优势,并利用了生物医学研究领域大量未被开发的文本资源。在统一的多模态特征空间中,BioMedGPT可灵活支持自由文本查询、处理多模态输入,并无需端到端微调即可赋能多种下游任务。
We open-source BioMedGPT-10B, a model instance of BioMedGPT, unifying texts, molecular structures, and protein sequences. BioMedGPT-10B is built upon BioMedGPT-LM-7B, which is fine-tuned from the recently released Llama2- Chat-7B model with millions of biomedical publications. We leverage independent encoders to encode molecular and protein features and project them into the same feature space of the textual modality via multi-modal fine-tuning. BioMedGPT-10B enables users to upload biological data of molecular structures and protein sequences and pose natural language queries about these data instances. This capability can potentially accelerate the discovery of novel molecular structures and protein functionalities, thus catalyzing advancements in drug development.
我们开源了BioMedGPT-10B,这是BioMedGPT的一个模型实例,统一了文本、分子结构和蛋白质序列。BioMedGPT-10B基于BioMedGPT-LM-7B构建,后者是从最近发布的Llama2-Chat-7B模型通过数百万篇生物医学文献微调而来。我们利用独立的编码器来编码分子和蛋白质特征,并通过多模态微调将它们投影到文本模态的同一特征空间中。BioMedGPT-10B允许用户上传分子结构和蛋白质序列的生物数据,并对这些数据实例提出自然语言查询。这一功能有望加速新型分子结构和蛋白质功能的发现,从而推动药物研发的进展。
Our contributions are summarized as follows:
我们的贡献总结如下:
• We introduce BioMedGPT, a novel framework to bridge the language of life and human natural language via large-scale generative language models. • We demonstrate the promising performance of BioMedGPT-10B, which is a model instance of BioMedGPT, on the biomedical QA, molecule QA, and protein QA tasks. Through fine-tuning, BioMedGPT-10B outperforms or is on par with human and significantly larger general-purpose foundation models on biomedical QA benchmarks. In addition, its capability on the molecule QA and protein QA tasks shows great potential to power many downstream biomedical applications, such as accelerating the discovery of new drugs and therapeutic targets. BioMedGPT-10B is open-sourced to the community. We also publish the datasets that are curated for the alignment of multi-modalities, i.e., PubChemQA and UniProtQA.
• 我们推出BioMedGPT,这是一个通过大规模生成式语言模型连接生命语言与人类自然语言的新颖框架。
• 我们展示了BioMedGPT-10B(BioMedGPT的一个模型实例)在生物医学问答、分子问答和蛋白质问答任务中的优异表现。通过微调,BioMedGPT-10B在生物医学问答基准测试中表现优于或与人类及规模显著更大的通用基础模型相当。此外,其在分子问答和蛋白质问答任务上的能力显示出推动众多下游生物医学应用的巨大潜力,例如加速新药和治疗靶点的发现。BioMedGPT-10B已向社区开源。我们还发布了为多模态对齐而整理的数据集,即PubChemQA和UniProtQA。
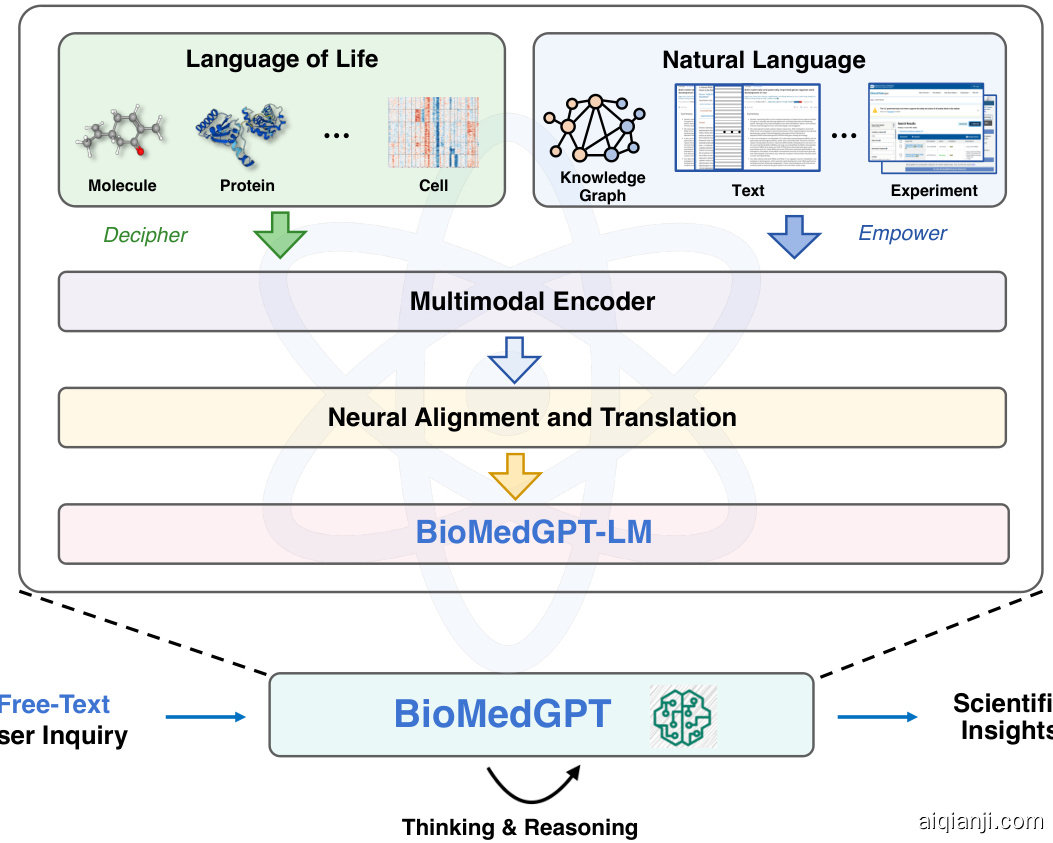
Figure 1: The architecture of BioMedGPT.
图 1: BioMedGPT 架构。
• BioMedGPT-LM-7B, the large language model used in BioMedGPT-10B, is the first generative language model that is fine-tuned from Llama2 with an extensive biomedical corpus. BioMedGPT-LM-7B is commercial friendly and is open-sourced to the community.
• BioMedGPT-LM-7B是BioMedGPT-10B中使用的大语言模型,也是首个基于Llama2并通过海量生物医学语料微调的生成式语言模型。该模型支持商业用途,并已向社区开源。
The remaining of the paper is organized as follows. Section 2 provides an overview of BioMedGPT. In Section 3, we present BioMedGPT-10B, which is a 10B foundation model in the BioMedGPT family, including BioMedGPT-LM-7B and molecule QA and protein QA modules. Experimental results and analysis are reported in Section 4. We describe the limitations of our work in Section 5. Finally, we conclude and discuss future work in Section 6.
论文的其余部分组织如下。第2节概述了BioMedGPT。第3节介绍了BioMedGPT-10B,这是BioMedGPT系列中的一个100亿参数基础模型,包括BioMedGPT-LM-7B以及分子问答和蛋白质问答模块。实验结果与分析在第4节中报告。我们在第5节描述了工作的局限性。最后,第6节总结并讨论了未来工作。
2 An Overview of BioMedGPT
2 BioMedGPT 概述
In Figure 1, we present an overview of BioMedGPT, which serves as a biomedical brain. The cognitive core, BioMedGPT-LM, is a large language model developed through incremental training on an extensive biomedical corpus, inheriting the benefits of both the emergent abilities of LLMs and domain-specific knowledge. BioMedGPT-LM serves not only as a linguistic engine that enables free-text interactions with humans but also acts as a bridge connecting various biomedical modalities. BioMedGPT is endowed with the ability to comprehend and reason over diverse biological modalities encompassing molecules, proteins, transcript omi c, and more, through the feature space alignment. In addition, we also utilize heterogeneous expert knowledge from knowledge graphs, text documents, and experimental results to further enhance the biomedical knowledge of BioMedGPT.
图1: BioMedGPT概览图,该系统作为生物医学大脑。其认知核心BioMedGPT-LM是通过海量生物医学语料增量训练开发的大语言模型,兼具大语言模型的涌现能力和领域专有知识优势。该模型不仅是支持人类自由文本交互的语言引擎,更是连接多模态生物医学数据的桥梁。通过特征空间对齐技术,BioMedGPT具备理解分子、蛋白质、转录组等多元生物模态并进行推理的能力。此外,我们还整合知识图谱、文本文档及实验结果等异构专家知识,进一步强化系统的生物医学知识体系。
We develop a feature fusion approach, which encodes multimodal data with independent pre-trained encoders and aligns their feature spaces with that of the natural language through neural alignment and translation methods. We have demonstrated the effectiveness of this approach in our prior work, MolFM [Luo et al., 2023], which is a molecular foundation model that enables joint representation learning on molecular structures, biomedical texts, and knowledge graphs. MolFM achieves state-of-the-art performance on a spectrum of multimodal tasks, such as molecule-text retrieval, molecule captioning, and text-to-molecule generation. Thus, we believe that BioMedGPT can harvest from both the readily available uni-modal foundation models and the powerful generalization capability of language models. BioMedGPT enhances the cross-modal comprehension and connections between natural language and diverse biological modalities. Users can flexibly present inquiries in various formats, encompassing but not limited to, text, chemical structure files, SMILES, protein sequences, protein 3D structure data, and single-cell sequencing data. With extensive training on these biomedical data, BioMedGPT can provide users with valuable scientific insights.
我们开发了一种特征融合方法,该方法通过独立的预训练编码器对多模态数据进行编码,并通过神经对齐和翻译方法将其特征空间与自然语言对齐。我们在先前的工作MolFM [Luo et al., 2023]中验证了该方法的有效性。MolFM是一个分子基础模型,能够对分子结构、生物医学文本和知识图谱进行联合表征学习,在分子-文本检索、分子描述生成和文本到分子生成等多模态任务上实现了最先进的性能。因此,我们相信BioMedGPT既能利用现成的单模态基础模型,又能继承大语言模型强大的泛化能力。BioMedGPT增强了自然语言与多种生物模态之间的跨模态理解和关联。用户可以灵活地以多种格式提交查询,包括但不限于文本、化学结构文件、SMILES、蛋白质序列、蛋白质3D结构数据和单细胞测序数据。通过对这些生物医学数据进行广泛训练,BioMedGPT能为用户提供有价值的科学洞见。
In the subsequent section, we introduce a model instance of BioMedGPT which is primarily focused on the joint comprehension of molecular structures, protein sequences, and biomedical texts.
在接下来的部分,我们将介绍BioMedGPT的一个模型实例,其主要关注分子结构、蛋白质序列和生物医学文本的联合理解。
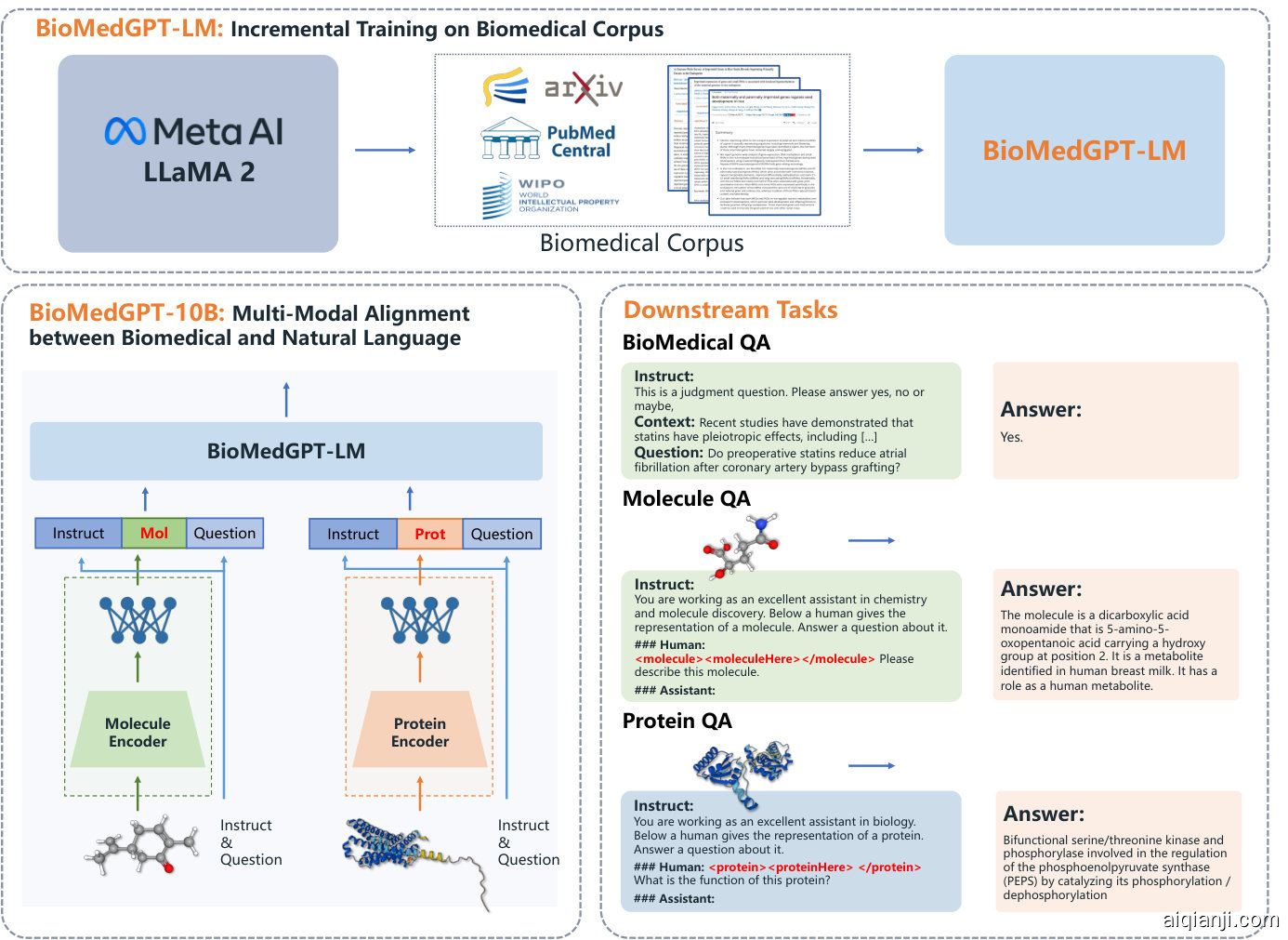
Figure 2: The overview of BioMedGPT-10B. BioMedGPT-LM is the large language model of BioMedGPT, which serves as a cognitive core to jointly comprehend various biological modalities through natural language. In BioMedGPT-10B, the parameter size of the large language model is 7B. BioMedGPT-10B adopts GraphMVP [Liu et al., 2022] as the 2D molecular graph encoder, ESM2-3B [Lin et al., 2022] as the protein sequence encoder, and conducts feature space alignment via a neural network adaptor. BioMedGPT can be applied to many multimodal downstream tasks such as biomedical QA, molecule QA, and protein QA.
图 2: BioMedGPT-10B 概述。BioMedGPT-LM 是 BioMedGPT 的大语言模型 (Large Language Model),作为认知核心通过自然语言联合理解多种生物模态。在 BioMedGPT-10B 中,大语言模型的参数量为 7B。BioMedGPT-10B 采用 GraphMVP [Liu et al., 2022] 作为 2D 分子图编码器,ESM2-3B [Lin et al., 2022] 作为蛋白质序列编码器,并通过神经网络适配器进行特征空间对齐。BioMedGPT 可应用于生物医学问答、分子问答和蛋白质问答等多模态下游任务。
3 BioMedGPT-10B: Aligning Molecules, Proteins, and Natural Language
3 BioMedGPT-10B:对齐分子、蛋白质与自然语言
As shown in Figure 2, we develop BioMedGPT-LM-7B, a large language model specialized in bio medicine, through incremental training with biomedical literature on top of Llama2-7B-Chat. We then build BioMedGPT-10B by aligning 2D molecular graphs, protein sequences, and natural language in a unified feature space. We choose molecules and proteins because they are basic biomedical elements. It is worth noting that the aforementioned molecule and protein encoders can be easily replaced with other suitable and well-performing encoders. We detail the architecture and training process of BioMedGPT-10B in the following subsections.
如图 2 所示, 我们在 Llama2-7B-Chat 的基础上通过生物医学文献的增量训练, 开发了专注于生物医学领域的大语言模型 BioMedGPT-LM-7B。随后通过将 2D 分子图、蛋白质序列和自然语言对齐到统一特征空间, 构建了 BioMedGPT-10B。选择分子和蛋白质是因为它们是基础的生物医学元素。值得注意的是, 上述分子和蛋白质编码器可以轻松替换为其他合适且性能良好的编码器。我们将在后续小节详细阐述 BioMedGPT-10B 的架构和训练过程。
3.1 BioMedGPT-LM-7B: Incremental training on large-scale biomedical literature
3.1 BioMedGPT-LM-7B:基于大规模生物医学文献的增量训练
Recently, Llama2 and Llama2-chat, open-sourced by Meta, have attracted great research attention owing to their outstanding capabilities in the general domain. To exploit the advantages of Llama2’s emergent abilities as well as biomedical knowledge from scientific research, we perform incremental training on Llama2-Chat-7B with extensive biomedical documents from S2ORC [Lo et al., 2020].
近期,Meta开源的Llama2和Llama2-chat凭借其在通用领域的出色能力引发了广泛研究关注。为结合Llama2的涌现能力优势与科研领域的生物医学知识,我们采用S2ORC [Lo et al., 2020] 的海量生物医学文献对Llama2-Chat-7B进行了增量训练。
Data Processing We select biomedical-related literature from S2ORC to fine-tune Llama2. The S2ORC dataset comprises an extensive collection of 81.1 million English academic papers spanning various disciplines. We meticulously extract 5.5 million biomedical papers from this dataset using PubMed Central (PMC)-ID and PubMed ID as criteria. After removing articles without full text and those with duplicate IDs, we attain a refined dataset of 4.2 million articles. Our subsequent processing involves the removal of author information, reference citations, and chart data from the main body of each article. The remaining text is then partitioned into sentence-based chunks. Each of these chunks is further tokenized by the Llama2 tokenizer, culminating in a substantial assemblage of over 26 billion tokens highly pertinent to the field of bio medicine.
数据处理
我们从S2ORC中选取生物医学相关文献对Llama2进行微调。S2ORC数据集包含跨越多个学科的8110万篇英文学术论文。我们以PubMed Central (PMC)-ID和PubMed ID为筛选标准,从该数据集中精细提取出550万篇生物医学论文。剔除无全文及ID重复的文章后,最终获得420万篇精炼数据集。后续处理包括移除每篇文章正文中的作者信息、参考文献引用及图表数据,剩余文本按句子分块后,通过Llama2的分词器(tokenizer)进一步切分,最终生成超过260亿个与生物医学领域高度相关的token。
Fine-tuning Details The fine-tuning utilizes a learning rate of $2\times10^{-5}$ , a batch size of $192^{3}$ , and a context length of 2048 tokens. We adopt the Fully Sharded Data Parallel (FSDP) acceleration strategy alongside the bf16 (Brain Floating Point) data format. To tackle the memory challenge, we leverage gradient check pointing [Chen et al., 2016] and flash attention [Dao et al., 2022]. We utilize an auto regressive objective function. The training loss is shown in Figure 3. Notably, the loss exhibits a consistent and progressive decrease after the 44,000 steps, indicating the model converges effectively.
微调细节
微调采用的学习率为 $2\times10^{-5}$,批量大小为 $192^{3}$,上下文长度为2048个token。我们采用全分片数据并行 (Fully Sharded Data Parallel, FSDP) 加速策略和bf16 (Brain Floating Point) 数据格式。为解决内存挑战,我们利用了梯度检查点 [Chen et al., 2016] 和闪存注意力机制 [Dao et al., 2022]。训练采用自回归目标函数,损失变化如图3所示。值得注意的是,在44,000步之后损失呈现持续稳定的下降趋势,表明模型有效收敛。

Figure 3: The training loss for BioMedGPT-LM-7B.
图 3: BioMedGPT-LM-7B 的训练损失。
3.2 Multimodal alignment between molecules, proteins, and natural language
3.2 分子、蛋白质与自然语言的多模态对齐
BioMedGPT-10B is composed of a molecule encoder, a protein encoder, a large language model (i.e., BioMedGPTLM-7B), and two modality adaptors. In order to exploit the strong capabilities of existing unimodal models, we leverage off-the-shelf checkpoints to initialize the molecule and protein encoder. The molecule encoder is a 5-layer GIN [Xu et al., 2018] with 1.8M parameters pre-trained by GraphMVP [Liu et al., 2022], which shows promising results in comprehending 2D molecular graphs. The protein encoder is ESM-2-3B [Lin et al., 2022], a 36-layer transformer [Vaswani et al., 2023] specialized in processing protein sequences. The output features of each atom of the molecule encoder and the output features of each residue of the protein encoder are projected to the feature space of BioMedGPT-LM-7B with independent modality adaptors composed of a fully-connected layer.
BioMedGPT-10B 由分子编码器、蛋白质编码器、大语言模型 (即 BioMedGPTLM-7B) 和两个模态适配器组成。为了利用现有单模态模型的强大能力,我们采用现成的检查点初始化分子和蛋白质编码器。分子编码器是一个具有 180 万参数的 5 层 GIN [Xu et al., 2018],通过 GraphMVP [Liu et al., 2022] 进行预训练,在理解 2D 分子图方面表现出色。蛋白质编码器是 ESM-2-3B [Lin et al., 2022],一个专用于处理蛋白质序列的 36 层 Transformer [Vaswani et al., 2023]。分子编码器每个原子的输出特征和蛋白质编码器每个残基的输出特征,通过由全连接层组成的独立模态适配器投影到 BioMedGPT-LM-7B 的特征空间。
To build the connections between molecular and protein structures with natural language, we perform multimodal fine-tuning, which involves answering questions with regard to a given molecule or protein. As shown in Table 1, we design prompt templates to help BioMedGPT-LM understand the context more accurately in a role-play manner. The
为了建立分子与蛋白质结构同自然语言之间的联系,我们进行了多模态微调,即针对给定分子或蛋白质回答问题。如表1所示,我们设计了提示模板,以角色扮演方式帮助BioMedGPT-LM更准确地理解上下文。其中
Data Proc ces sing The multimodal alignment is performed on two large-scale datasets that we curate, namely, PubChemQA and UniProtQA. We publicly release these datasets to facilitate future research.
数据处理
我们在两个自建的大规模数据集PubChemQA和UniProtQA上执行多模态对齐任务。这些数据集已公开发布以促进后续研究。
• PubChemQA consists of molecules and their corresponding textual descriptions from PubChem [Kim et al., 2022]. It contains a single type of question, i.e., please describe the molecule. We remove molecules that cannot be processed by RDKit [Landrum et al., 2021] to generate 2D molecular graphs. We also remove texts with less than 4 words, and crops descriptions with more than 256 words. Finally, we obtain 325, 754 unique molecules and 365, 129 molecule-text pairs. On average, each text description contains 17 words. • UniProtQA consists of proteins and textual queries about their functions and properties. The dataset is constructed from UniProt [Consortium, 2022], and consists 4 types of questions with regard to functions, official names, protein families, and sub-cellular locations. We collect a total of 569, 516 proteins and 1, 891, 506 question-answering samples. The data was randomly divided into training, validation, and test sets at a ratio of $8:1:1$ . The multi-modal fine-tuning is performed on the training set of ProteinQA.
• PubChemQA 包含来自 PubChem [Kim et al., 2022] 的分子及其对应的文本描述。它仅包含一种问题类型,即"请描述该分子"。我们移除了无法通过 RDKit [Landrum et al., 2021] 处理以生成二维分子图的分子,同时删除了少于4个单词的文本,并将超过256个单词的描述进行截断。最终获得325,754个独特分子和365,129个分子-文本对。平均每个文本描述包含17个单词。
• UniProtQA 包含蛋白质及其功能和特性的文本查询。该数据集构建自 UniProt [Consortium, 2022],包含关于功能、官方名称、蛋白质家族和亚细胞定位的4类问题。我们共收集了569,516个蛋白质和1,891,506个问答样本。数据按 $8:1:1$ 的比例随机划分为训练集、验证集和测试集。多模态微调在 ProteinQA 的训练集上进行。
We design the following prompt to organize the molecular or protein data with the text data as feature-ordered input for the LLM.
我们设计了以下提示(prompt),用于将分子或蛋白质数据与文本数据整理为特征有序的输入,供大语言模型使用。
Table 1: Prompt for organizing multi-modality data entry.
| Modality | Prompt |
| Molecule | You are working as an excellent assistant in chemistry and molecule discovery. Below a human gives the representation of a molecule. Answer a question about it. ###Human: |
| Protein | ###Assistant: {text_ output} You are working as an excellent assistant in biology.Below a human gives the representation of a protein. Answer a question about it. ###Human: |
表 1: 多模态数据录入的组织提示
| 模态类型 | 提示语 |
|---|---|
| 分子 (Molecule) | 你是一名优秀的化学与分子发现助手。以下用户提供了一个分子表示,请回答相关问题。 ###Human: {text_input}。 |
| 蛋白质 (Protein) | ###Assistant: {text_ output} 你是一名优秀的生物学助手。以下用户提供了一个蛋白质表示,请回答相关问题。 ###Human: {text_input}。 ###Assistant: {text_ output} |
Following mPLUG-owl [Ye et al., 2023], we freeze the parameters of BioMedGPT-LM and optimize the parameters of the molecule encoder, protein encoder, and modality adaptors to save the computational cost and avoid catastrophic forgetting. We conduct fine-tuning using these two datasets.
遵循mPLUG-owl [Ye et al., 2023]的方法,我们冻结了BioMedGPT-LM的参数,并优化分子编码器、蛋白质编码器和模态适配器的参数,以节省计算成本并避免灾难性遗忘。我们使用这两个数据集进行微调。
4 Experiment
4 实验
In this section, we substantiate BioMedGPT-10B’s capability to jointly understand and model the language of life and natural language with a series of experiments. We present three question-answering tasks, namely biomedical QA, molecule QA and protein QA, to comprehensively evaluate the biomedical knowledge that our model encompasses. In the following sections, we will introduce the datasets and experimental results for each task. Additionally, we showcase the generated results in protein QA.
在本节中,我们通过一系列实验验证了BioMedGPT-10B联合理解与建模生命语言和自然语言的能力。我们设计了三个问答任务(即生物医学QA、分子QA和蛋白质QA)来全面评估模型所涵盖的生物医学知识。后续章节将分别介绍各任务的数据集与实验结果,并展示蛋白质QA的生成案例。
4.1 Biomedical QA
4.1 生物医学问答
Biomedical QA involves answering free-text questions in the biomedical domain, which challenges the professional level of language models. The task serves as a means to evaluate if BioMedGPT-10B can understand biomedical terminologies and reason over complex contexts like a human expert.
生物医学问答涉及回答生物医学领域的自由文本问题,这对语言模型的专业水平提出了挑战。该任务旨在评估BioMedGPT-10B是否能像人类专家
