LLaVA-Med: Training a Large Language-and-Vision Assistant for Bio medicine in One Day
LLaVA-Med: 一天内训练出用于生物医学的大语言视觉助手
Chunyuan $\mathbf{Li}^{* }$ , Cliff Wong∗, Sheng Zhang∗, Naoto Usuyama, Haotian Liu, Jianwei Yang Tristan Naumann, Hoifung Poon, Jianfeng Gao
Chunyuan $\mathbf{Li}^{* }$, Cliff Wong∗, Sheng Zhang∗, Naoto Usuyama, Haotian Liu, Jianwei Yang Tristan Naumann, Hoifung Poon, Jianfeng Gao
Microsoft https://aka.ms/llava-med
Microsoft https://aka.ms/llava-med
Abstract
摘要
Conversational generative AI has demonstrated remarkable promise for empowering biomedical practitioners, but current investigations focus on unimodal text. Multimodal conversational AI has seen rapid progress by leveraging billions of image-text pairs from the public web, but such general-domain vision-language models still lack sophistication in understanding and conversing about biomedical images. In this paper, we propose a cost-efficient approach for training a visionlanguage conversational assistant that can answer open-ended research questions of biomedical images. The key idea is to leverage a large-scale, broad-coverage biomedical figure-caption dataset extracted from PubMed Central, use GPT-4 to self-instruct open-ended instruction-following data from the captions, and then fine-tune a large general-domain vision-language model using a novel curriculum learning method. Specifically, the model first learns to align biomedical vocabulary using the figure-caption pairs as is, then learns to master open-ended conversational semantics using GPT-4 generated instruction-following data, broadly mimicking how a layperson gradually acquires biomedical knowledge. This enables us to train a Large Language and Vision Assistant for Bio Medicine (LLaVA-Med) in less than 15 hours (with eight A100s). LLaVA-Med exhibits excellent multimodal convers at ional capability and can follow open-ended instruction to assist with inquiries about a biomedical image. On three standard biomedical visual question answering datasets, fine-tuning LLaVA-Med outperforms previous supervised state-of-the-art on certain metrics. To facilitate biomedical multimodal research, we will release our instruction-following data and the LLaVA-Med model.
对话式生成式AI(Generative AI)在赋能生物医学实践方面展现出巨大潜力,但现有研究主要集中于单模态文本。通过利用公共网络的数十亿图文对,多模态对话式AI已取得快速进展,然而这类通用领域视觉语言模型在理解和讨论生物医学图像方面仍显不足。本文提出一种高效训练视觉语言对话助手的方法,使其能够回答生物医学图像的开放式研究问题。核心思路是从PubMed Central提取大规模、广覆盖的生物医学图文数据集,利用GPT-4从图注自动生成开放式指令遵循数据,再通过新颖的课程学习方式微调通用领域视觉语言大模型。具体而言,模型先通过原始图文对学习生物医学术语对齐,再通过GPT-4生成的指令数据掌握开放式对话语义,模拟普通人逐步获取生物医学知识的过程。该方法仅需不到15小时(使用8块A100)即可训练出生物医学大语言视觉助手(LLaVA-Med)。LLaVA-Med展现出卓越的多模态对话能力,能遵循开放式指令协助解析生物医学图像。在三个标准生物医学视觉问答数据集上,微调后的LLaVA-Med在部分指标上超越了此前有监督的最先进方法。为促进生物医学多模态研究,我们将公开指令数据集和LLaVA-Med模型。
1 Introduction
1 引言
Parallel image-text data is abundantly available in the general domain, such as web images and their associated captions. Generative pre training has proven effective to leverage this parallel data for self-supervised vision-language modeling, as demonstrated by multimodal GPT-4 [32] and opensourced efforts such as LLaVA [24]. By instruction-tuning models to align with human intents based on multimodal inputs, the resulting large multimodal models (LMMs) exhibit strong zero-shot task completion performance on a variety of user-oriented vision-language tasks such as image understanding and reasoning, paving the way to develop general-purpose multimodal conversational assistants [2, 21, 9].
通用领域存在大量并行的图文数据,例如网络图片及其关联标题。生成式预训练 (Generative Pre-training) 已被证明能有效利用这种并行数据进行自监督视觉语言建模,多模态GPT-4 [32] 和开源项目LLaVA [24] 等成果已验证了这一点。通过基于多模态输入对模型进行指令微调 (instruction-tuning) 以对齐人类意图,最终得到的大规模多模态模型 (LMMs) 在面向用户的视觉语言任务(如图像理解与推理)中展现出强大的零样本任务完成能力,为开发通用多模态对话助手铺平了道路 [2, 21, 9]。
While successful in the general domains, such LMMs are less effective for biomedical scenarios because biomedical image-text pairs are drastically different from general web content. As a result, general-domain visual assistants may behave like a layperson, who would refrain from answering biomedical questions, or worse, produce incorrect responses or complete hallucinations. Much progress has been made in biomedical visual question answering (VQA), but prior methods typically formulate the problem as classification (e.g., among distinct answers observed in the training set) and are not well equipped for open-ended instruction-following. Consequently, although conversational generative AI has demonstrated great potential for biomedical applications [19, 30, 18], current investigations are often limited to unimodal text.
虽然在通用领域取得了成功,但此类大语言模型在生物医学场景中的效果较差,因为生物医学图文对与通用网络内容截然不同。因此,通用领域的视觉助手可能表现得像外行,要么回避回答生物医学问题,要么更糟,产生错误响应或完全幻觉。生物医学视觉问答 (VQA) 已取得很大进展,但现有方法通常将问题表述为分类任务 (例如在训练集中观察到的不同答案之间进行选择),并不擅长处理开放式指令跟随。因此,尽管对话式生成式 AI (Generative AI) 在生物医学应用中展现出巨大潜力 [19, 30, 18],但目前的研究往往局限于单模态文本。
In this paper, we present Large Language and Vision Assistant for Bio Medicine (LLaVA-Med), a first attempt to extend multimodal instruction-tuning to the biomedical domain for end-to-end training of a biomedical multimodal conversational assistant. Domain-specific pre training has been shown to be effective for biomedical natural language processing (NLP) applications [17, 14, 10, 28] and biomedical vision-language (VL) tasks [15, 7, 38, 49, 8]. Most recently, large-scale biomedical VL learning has been made possible by the creation of PMC-15M [49], a broad-coverage dataset with 15 million biomedical image-text pairs extracted from PubMed Central1. This dataset is two orders of magnitude larger than the next largest public dataset, MIMIC-CXR [15], and covers a diverse image types. Inspired by recent work in instruction-tuning [34, 24], LLaVA-Med uses GPT-4 to generate diverse biomedical multimodal instruction-following data using image-text pairs from PMC-15M, and fine-tune a large biomedical-domain VL model [24] using a novel curriculum learning method.
本文介绍了生物医学领域的大语言与视觉助手(LLaVA-Med),这是将多模态指令调优技术扩展到生物医学领域、实现端到端生物医学多模态对话助手训练的首次尝试。领域特异性预训练已被证明对生物医学自然语言处理(NLP)应用[17,14,10,28]和生物医学视觉语言(VL)任务[15,7,38,49,8]具有显著效果。最近,通过创建PMC-15M[49]这一包含1500万对生物医学图文数据的大型数据集,使得大规模生物医学VL学习成为可能。该数据集规模比第二大公开数据集MIMIC-CXR[15]大两个数量级,且涵盖多样化的图像类型。受指令调优领域最新研究[34,24]启发,LLaVA-Med利用GPT-4基于PMC-15M的图文对生成多样化生物医学多模态指令跟随数据,并采用新颖课程学习方法对大型生物医学领域VL模型[24]进行微调。
Specifically, our paper makes the following contributions:
具体而言,本文的贡献如下:
2 Related Work
2 相关工作
Biomedical Chatbots. Inspired by ChatGPT [31]/GPT-4 [32] and the success of open-sourced instruction-tuned large language models (LLMs) in the general domain, several biomedical LLM chatbots have been developed, including ChatDoctor [47], Med-Alpaca [12], PMC-LLaMA [45], Clinical Camel [1], DoctorGLM [46], and Huatuo [44]. They are initialized with open-sourced LLM and fine-tuned on customized sets of biomedical instruction-following data. The resulting LLMs emerge with great potential to offer assistance in a variety of biomedical-related fields/settings, such as understanding patients’ needs and providing informed advice.
生物医学聊天机器人。受ChatGPT [31]/GPT-4 [32]以及开源指令调优大语言模型(LLM)在通用领域成功的启发,目前已开发出多款生物医学LLM聊天机器人,包括ChatDoctor [47]、Med-Alpaca [12]、PMC-LLaMA [45]、Clinical Camel [1]、DoctorGLM [46]和华佗(Huatuo) [44]。这些模型基于开源LLM初始化,并在定制化的生物医学指令遵循数据集上进行微调。最终形成的LLM展现出在多种生物医学相关领域/场景中提供协助的巨大潜力,例如理解患者需求并提供专业建议。
To our knowledge, Visual Med-Alpaca [39] is the only existing multimodal biomedical chatbot that accepts image inputs. Though Visual Med-Alpaca and the proposed LLaVA-Med share a similar input-output data format, they differ in key aspects: (i) Model architectures. LLaVA-Med is an end-to-end neural model and Visual Med-Alpaca is a system that connect multiple image captioning models with a LLM, using a classifier to determine if or which biomedical captioning model is responsible for the image. The text prompt subsequently merges the converted visual information with the textual query, enabling Med-Alpaca to generate an appropriate response. $(i i)$ Biomedical instruction-following data. While Visual Med-Alpaca is trained on 54K samples from limited biomedical subject domains, LLaVA-Med is trained a more diverse set.
据我们所知,Visual Med-Alpaca [39] 是目前唯一支持图像输入的多模态生物医学对话系统。尽管Visual Med-Alpaca与本文提出的LLaVA-Med具有相似的输入输出数据格式,但二者存在关键差异:(i) 模型架构。LLaVA-Med是端到端神经网络模型,而Visual Med-Alpaca是通过分类器判断图像归属、串联多个图像描述模型与大语言模型的系统,其文本提示会将转换后的视觉信息与文本查询合并,使Med-Alpaca生成响应。$(ii)$ 生物医学指令数据。Visual Med-Alpaca仅基于54K有限生物医学领域样本训练,而LLaVA-Med采用了更丰富多样的训练集。
Biomedical Visual Question Answering. An automated approach to building models that can answer questions based on biomedical images stands to support clinicians and patients. To describe existing biomedical VQA methods, we make a distinction between disc rim i native and generative methods. For disc rim i native methods, VQA is treated a classification problem: models make predictions from a predefined set of answers. While disc rim i native methods yield good performance, they deal with closed-set predictions [13], and require mitigation when a customized answer set is provided in at inference [22, 49, 8]. The disc rim i native formulation is suboptimal towards the goal of developing a general-purpose biomedical assistant that can answer open questions in the wild. To this end, generative methods have been developed to predict answers as a free-form text sequence [5, 26, 41]. Generative methods are more versatile because they naturally cast the close-set questions as as special case where candidate answers are in language instructions.
生物医学视觉问答。构建能基于生物医学图像回答问题的自动化模型方法,有望为临床医生和患者提供支持。为描述现有生物医学VQA方法,我们区分了判别式(discriminative)与生成式(generative)方法。对于判别式方法,VQA被视为分类问题:模型从预定义答案集中进行预测。虽然判别式方法表现良好,但其处理的是封闭集预测[13],在推理时提供定制化答案集时需要额外处理[22,49,8]。这种判别式框架对于开发能回答开放问题的通用生物医学助手目标而言并非最优。为此,研究者开发了生成式方法,将答案预测为自由文本序列[5,26,41]。生成式方法更具通用性,因为它们天然将封闭集问题视为语言指令中包含候选答案的特殊情况。
Model Architecture. LLaVA-Med is similar to prefix tuning of language models (LMs) in [41] in that a new trainable module connects frozen image encoder and causal LM. In [41], a three-layer MLP network is used to map the visual features into a visual prefix, and the pre-trained LM are GPT2-XL [37], BioMedLM [42] and BioGPT [28], with size varying from 1.5B to 2.7B. By contrast, LLaVA-Med uses a linear projection and a 7B LM [43, 40]. Most importantly, [41] only considers standard supervised fine-tuning and focuses efforts on exploring various modeling choices. Our main contributions instead comprise proposing a novel data generation method that uses GPT-4 to self-instruct biomedical multimodal instruction-following data using freely-available broad-coverage biomedical image-text pairs extracted from PubMed Central [49].
模型架构。LLaVA-Med 类似于 [41] 中语言模型 (LM) 的前缀调优方法,通过一个新的可训练模块连接冻结的图像编码器和因果 LM。在 [41] 中,使用了一个三层 MLP 网络将视觉特征映射为视觉前缀,预训练的 LM 包括 GPT2-XL [37]、BioMedLM [42] 和 BioGPT [28],模型大小从 1.5B 到 2.7B 不等。相比之下,LLaVA-Med 使用了线性投影和一个 7B 的 LM [43, 40]。最重要的是,[41] 仅考虑了标准的有监督微调,并将重点放在探索各种建模选择上。而我们的主要贡献在于提出了一种新颖的数据生成方法,利用 GPT-4 通过自指令方式生成生物医学多模态指令跟随数据,这些数据基于从 PubMed Central [49] 提取的广泛覆盖的生物医学图文对。
3 Biomedical Visual Instruction-Following Data
3 生物医学视觉指令跟随数据
There are a lack of multimodal biomedical datasets to train an instruction-following assistant. To fill this gap, we create the first dataset of its kind from widely existing biomedical image-text pairs, through a machine-human co-curation procedure. It consists of two sets, concept alignment and instruction-following, which are used at different training stages, described in Section 4.
缺乏用于训练遵循指令助手的多模态生物医学数据集。为填补这一空白,我们通过机器-人工协同筛选流程,从广泛存在的生物医学图文对中创建了首个此类数据集。该数据集包含概念对齐和遵循指令两个子集,分别用于不同训练阶段(详见第4节)。
Biomedical Concept Alignment Data. For a biomedical image $\mathbf{X}_ {\mathrm{v}}$ and its associated caption $\mathbf{X}_ {\mathtt{c}}$ , we sample a question $\mathbf{X}_ {\mathfrak{q}}$ , which asks to describe the biomedical image. With $(\mathbf{X}_ {\mathtt{v}},\mathbf{X}_ {\mathtt{c}},\mathbf{X}_ {\mathtt{q}})$ , we create a single-round instruction-following example:
生物医学概念对齐数据。对于一张生物医学图像 $\mathbf{X}_ {\mathrm{v}}$ 及其关联标题 $\mathbf{X}_ {\mathtt{c}}$,我们采样一个问题 $\mathbf{X}_ {\mathfrak{q}}$,要求描述该生物医学图像。基于 $(\mathbf{X}_ {\mathtt{v}},\mathbf{X}_ {\mathtt{c}},\mathbf{X}_ {\mathtt{q}})$,我们构建一个单轮指令跟随示例:
$$
\mathrm{Human:X_ {q}X_ {v}{<}S T O P{>}\backslash n A s s i s t a n t:X_ {c}{<}S T O P{>}\backslash n}
$$
$$
\mathrm{Human:X_ {q}X_ {v}{<}S T O P{>}\backslash n A s s i s t a n t:X_ {c}{<}S T O P{>}\backslash n}
$$
Depending on the length of caption, the question that is sampled either asks to describe the image concisely or in detail. Two lists of questions are provided in Appendix A. In practice, $25%$ of captions have length less than 30 words in PMC-15M [49], and thus 30 words is used as the cutoff point to determine which list to choose. We sample 600K image-text pairs from PMC-15M. Though this dataset only presents one-single task instructions, i.e., image captioning, it contains a diverse and representative set of biomedical concept samples from the original PMC-15M [49].
根据标题的长度,采样到的问题会要求简洁或详细地描述图像。附录A中提供了两个问题列表。实际上,在PMC-15M [49]中,25%的标题长度少于30个词,因此以30词作为选择问题列表的分界点。我们从PMC-15M中采样了60万张图像-文本对。尽管该数据集仅包含单一任务指令(即图像描述),但它从原始PMC-15M [49]中提取了多样化且具有代表性的生物医学概念样本。
Biomedical Instruction-Tuning Data. To align the model to follow a variety of instructions, we present and curate diverse instruction-following data with multi-round conversations about the provided biomedical images, by prompting language-only GPT-4. Specifically, given an image caption, we design instructions in a prompt that asks GPT-4 to generate multi-round questions and answers in a tone as if it could see the image (even though it only has access to the text). Sometimes the image caption is too short for GPT-4 to generate meaningful questions and answers. To provide more context regarding the image, we also create a prompt that includes not only captions but also sentences from the original PubMed paper that mentions the image. We also manually curate few-shot examples in the prompt to demonstrate how to generate high-quality conversations based on the provided caption and context. See Appendix B for the prompt and few-shot examples. To collect image captions and their context, we filter PMC-15M to retain the images that only contain a single plot. From them, we sample 60K image-text pairs from the five most common imaging modalities: CXR (chest X-ray), CT (computed tomography), MRI (magnetic resonance imaging), his to pathology, and gross (i.e., macroscopic) pathology. We then extract sentences that mention the image from the original PubMed paper as additional context to the caption, inspired by the observations that external knowledge helps generalization [20, 25].
生物医学指令微调数据。为使模型能够遵循多样化指令,我们通过仅使用文本的GPT-4生成多轮对话,整理出关于给定生物医学图像的多样化指令跟随数据。具体而言,给定图像描述时,我们设计提示词要求GPT-4以仿佛能看到图像的语调生成多轮问答(尽管其仅能访问文本)。当图像描述过短时,我们会创建同时包含描述和提及该图像的PubMed论文句子的提示词以提供更多上下文。我们还手动编写了提示词中的少样本示例,展示如何基于给定描述和上下文生成高质量对话(详见附录B的提示词和少样本示例)。为收集图像描述及其上下文,我们从PMC-15M筛选仅含单个图表图像,从中按五种最常见成像模态(胸透X光(CXR)、计算机断层扫描(CT)、磁共振成像(MRI)、组织病理学和大体病理学)抽取6万图文对,并借鉴外部知识提升泛化能力的研究[20,25],从原始PubMed论文提取提及图像的句子作为描述的补充上下文。
An example of instruction-following data is shown in Figure 1 shows, and the data statistics is shown Figure 2. We have produced three versions of instruct data when iterative ly improving the data
指令跟随数据的示例如图1所示,数据统计信息如图2所示。我们在迭代改进数据时生成了三个版本的指令数据。
Biomedical Visual Instruction-Following Example
生物医学视觉指令跟随示例
Caption:
标题:
Figure 4: Contrast-enhanced CT scan of the chest for patient #1. A large cavitating lesion was identified in the right upper lobe with pleural involvement and likely extension into the chest wall. In addition, right hilar lymph aden opa thy was suspected. This was the primary malignant lesion in this patient.
图 4: 患者#1的胸部增强CT扫描。右肺上叶发现一个大型空洞性病变,累及胸膜并可能延伸至胸壁。此外,怀疑存在右肺门淋巴结病变。这是该患者的主要恶性病灶。
Sentences with in-line figure mentions:
带内联图表引用的句子:


- Computerized tomography ( CT ) scans of the chest ( Figure 4 ) were obtained. - ( Figure 4 ) and demonstrated a large cavitating lesion in the posterior aspect of the right upper lobe with probable pleural involvement likely representing the primary lesion.
- 进行了胸部计算机断层扫描 (CT) (图 4: )
- (图 4: ) 显示右肺上叶后部有一个大型空洞性病变,可能累及胸膜,很可能代表原发病灶。
Conversation:
对话:
User: Where is the lesion located?
病灶位于何处?
Assistant: The lesion is located in the posterior aspect of the right upper lobe of the lung.
病变位于右肺上叶后部。
User: Is it extending into the chest wall?
用户:是否已扩散至胸壁?
Assistant: Yes, the CT scan suggests that the large cavitating lesion in the right upper lobe is likely extending into the chest wall.
CT扫描提示右肺上叶的大空洞性病变可能已侵犯胸壁。
User: Can you explain more about the cavitating lesion?
用户: 能详细解释一下空泡性病变吗?
Assistant: Certainly. A cavitating lesion refers to an abnormal mass or growth within the lung tissue that has a cavity or hollow space inside it. This cavity can be formed due to the necrosis (death) of the tissue inside the lesion, leading to the breakdown and disintegration of the affected tissue. Cavitating lesions can be caused by various factors, such as infections (like tuberculosis or fungal infections), autoimmune diseases, or malignancies (cancer). In the context of the CT scan provided, the large cavitating lesion in the right upper lobe of the lung is suggestive of a malignant (cancerous) growth. The cavity within the lesion may be a result of the rapid and uncontrolled growth of cancer cells, which can lead to the destruction of surrounding healthy tissue. It’s important to note that further diagnostic tests, such as a biopsy, would be required to confirm the malignancy and determine the specific type of cancer involved.
规则:
- 输出中文翻译部分时,仅保留翻译标题,不包含任何冗余内容、重复或解释。
- 不输出与英文无关的内容。
- 保留原始段落格式及术语(如FLAC、JPEG)、公司缩写(如Microsoft、Amazon、OpenAI)。
- 人名不翻译。
- 保留论文引用格式(如[20])。
- Figure/Table翻译为“图/表”并保留编号(如“图1: ”、“表1: ”)。
- 全角括号替换为半角括号,左右括号前后添加半角空格。
- 专业术语首次出现时标注英文(如“生成式AI (Generative AI)”),后续仅用中文。
策略:
- 特殊字符/公式原样保留
- HTML表格转为Markdown格式
- 翻译内容需符合中文表达习惯且无信息遗漏
最终仅返回Markdown格式译文。
-
- 翻译结果:* *
空泡性病变指肺组织内存在空腔或中空结构的异常肿块。该空腔可能由病变内部组织坏死(组织死亡)导致,引发受累组织的分解与破坏。空泡性病变的成因多样,包括感染(如结核或真菌感染)、自身免疫性疾病或恶性肿瘤(癌症)。根据提供的CT扫描结果,右肺上叶的大型空泡性病变提示恶性(癌性)肿块。病变内的空腔可能是癌细胞快速无序增殖破坏周围健康组织所致。需注意,需通过活检等进一步诊断确认恶性程度及具体癌症类型。
Figure 1: An instance of our GPT-4 generated instruction-following data. Top: The figure and caption were extracted from a PubMed Central full-text article [35], along with the corresponding citances (mentions of the given figure in the article). Bottom: The instruction-following data generated by GPT-4 using the text only (caption and citances). Note that the image is not used to prompt GPT-4; we only show it here as a reference.
图 1: 我们的GPT-4生成的指令遵循数据示例。顶部:该图和标题是从PubMed Central全文文章[35]中提取的,同时包含相应的引用(文章中对该图的提及)。底部:GPT-4仅使用文本(标题和引用)生成的指令遵循数据。请注意,图像并未用于提示GPT-4;我们在此仅将其作为参考展示。
quality: (i) 60K-IM. The a foremen i one d dataset that considers inline mentions (IM) as the context. (ii) 60K. A dataset of similar size (60K samples) without IM in self-instruct generation. (iii) 10K. A smaller dataset (10 samples) without IM. They are used to ablate our data generation strategies and their impact on trained LLaVA-Med in experiments.
质量:(i) 60K-IM。该数据集将内联提及(IM)作为上下文。(ii) 60K。一个规模相似(6万样本)但不含IM的自指令生成数据集。(iii) 10K。不含IM的小型数据集(10个样本)。这些数据集用于实验中消融研究我们的数据生成策略及其对训练的LLaVA-Med模型的影响。
4 Adapting Multimodal Conversational Models to the Biomedical Domain
4 将多模态对话模型适配到生物医学领域
We employ LLaVA,a general-domain multimodal conversation model [24], as the initial generaldomain LM, and continuously train the model to the biomedical domain. The same network architecture is utilized, where a linear projection layer connects the vision encoder and the language model. For LLaVA-Med model training, we use a two-stage procedure, illustrated in Figure 3.
我们采用LLaVA(一种通用领域多模态对话模型[24])作为初始通用领域大语言模型,并持续训练该模型至生物医学领域。该方案沿用相同网络架构,即通过线性投影层连接视觉编码器与语言模型。LLaVA-Med模型训练采用图3所示的两阶段流程:
Stage 1: Biomedical Concept Feature Alignment. To balance between concept coverage and training efficiency, we filter PMC-15M to 600K image-text pairs. These pairs are converted to instruction-following data using a naive expansion method: instructions simply presents the task of describing the image. For each sample, given the language instruction and image input, we ask the model to predict the original caption. In training, we keep both the visual encoder and LM weights frozen, and only update the projection matrix. In this way, the image features of vast novel biomedical visual concepts can be aligned to their textual word embeddings in the pre-trained LM. This stage can be understood as expanding the vocabulary of aligned image-text tokens to the biomedical domain.
阶段1:生物医学概念特征对齐。为平衡概念覆盖范围与训练效率,我们从PMC-15M数据集中筛选出60万张图文对。这些配对通过基础扩展方法转化为指令跟随数据:指令仅描述图像内容生成任务。针对每个样本,在给定语言指令和图像输入后,要求模型预测原始标题。训练过程中保持视觉编码器和大语言模型权重冻结,仅更新投影矩阵。通过这种方式,大量新型生物医学视觉概念的特征可与预训练大语言模型中的文本词向量对齐。该阶段可理解为将已对齐的图像-文本token词汇表扩展至生物医学领域。
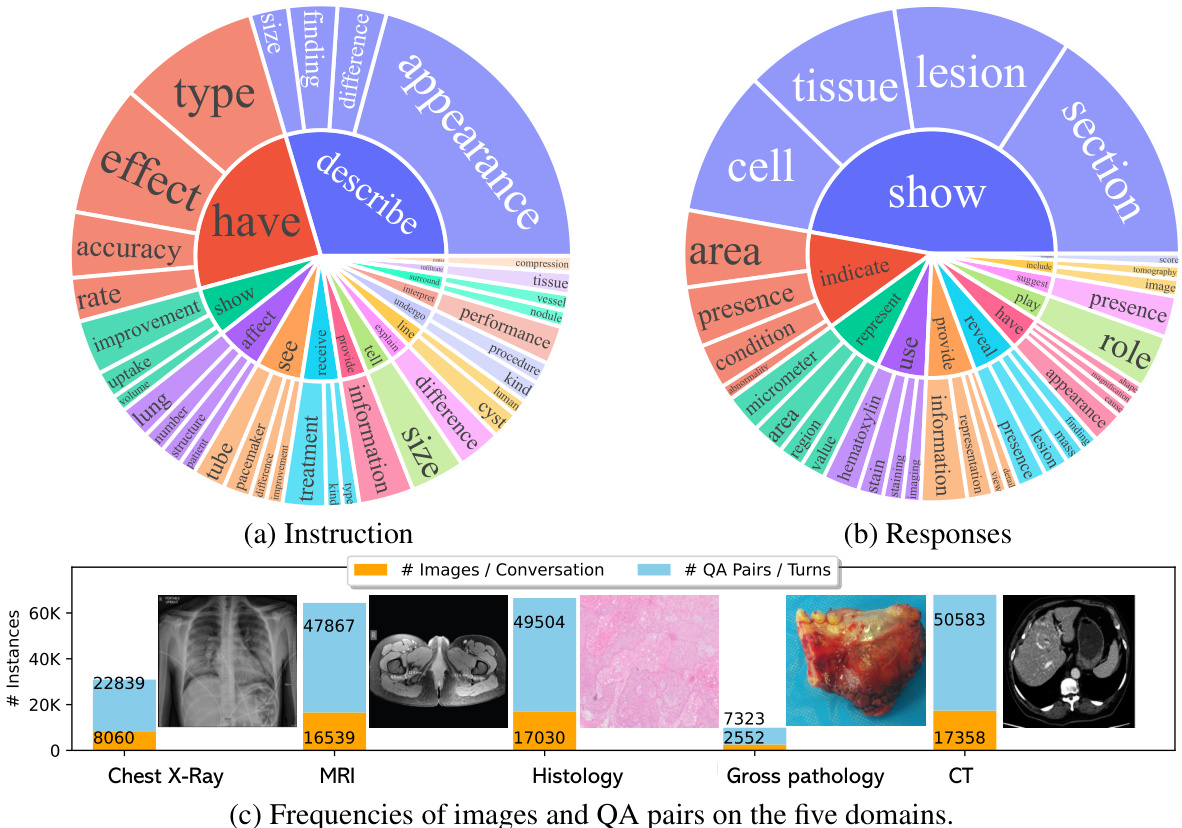
Figure 2: The data statistics of biomedical multimodal instruction-following data: (a,b) The root verb-noun pairs of instruction and responses, where the inner circle of the plot represents the root verb of the output response, and the outer circle represents the direct nouns. (c) The distribution of images and QA pairs on the five domains, one image is shown per domain. The domain example images are from [3, 33, 4, 29, 48].
图 2: 生物医学多模态指令跟随数据统计: (a,b) 指令与响应的根动词-名词对,其中图表内圈表示输出响应的根动词,外圈表示直接宾语名词。 (c) 五个领域上图像和问答对的分布,每个领域展示一张示例图像。领域示例图像来自 [3, 33, 4, 29, 48]。
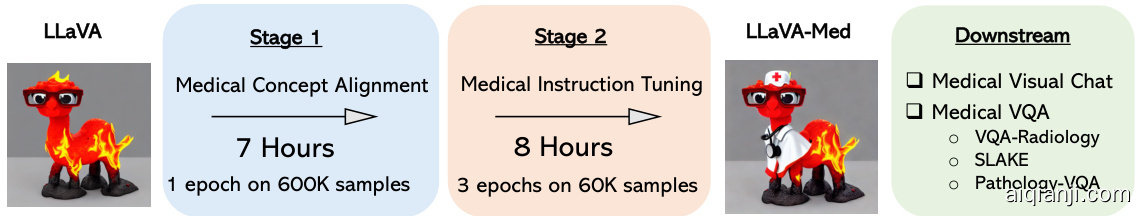
Figure 3: LLaVA-Med was initialized with the general-domain LLaVA and then continuously trained in a curriculum learning fashion (first biomedical concept alignment then full-blown instructiontuning). We evaluated LLaVA-Med on standard visual conversation and question answering tasks.
图 3: LLaVA-Med 基于通用领域 LLaVA 初始化,随后采用课程学习方式进行持续训练 (先进行生物医学概念对齐,再进行完整指令微调)。我们在标准视觉对话和问答任务上评估了 LLaVA-Med。
Stage 2: End-to-End Instruction-Tuning. We only keep the visual encoder weights frozen, and continue to update both the pre-trained weights of the projection layer and LM. To train the model to follow various instructions and complete tasks in a conversational manner, we develop a biomedical chatbot by fine-tuning our model on the biomedical language-image instruction-following data collected in Section 3. As demonstrated in the experiments to be described later, the LLaVA-Med model at this stage is able to not only be served as a biomedical visual assistant to interact with users, but also achieve good zero-shot task transfer performance when evaluated on well-established biomedical VQA datasets.
阶段2:端到端指令微调。我们仅冻结视觉编码器的权重,继续更新投影层和大语言模型的预训练权重。为了让模型学会以对话方式遵循各类指令并完成任务,我们在第3节收集的生物医学图文指令跟随数据上微调模型,开发出生物医学聊天机器人。如后续实验所示,此阶段的LLaVA-Med模型不仅能作为生物医学视觉助手与用户交互,在成熟的生物医学VQA数据集上进行零样本任务迁移评估时也表现出色。
Fine-tuning to Downstream Datasets. For some specific biomedical scenarios, there is a need of developing highly accurate and dataset-specific models to improve the service quality of the assistant. We fine-tune LLaVA-Med after the two-stage training on three biomedical VQA datasets [27], covering varied dataset sizes and diverse biomedical subjects. Given a biomedical image as context, multiple natural language questions are provided, the assistant responds in free-form text for both the close-set and open-set questions, with a list of candidate answers constructed in the prompt for each close-set question.
针对下游数据集的微调。在某些特定生物医学场景中,需要开发高精度且针对特定数据集的模型以提升助手服务质量。我们在两阶段训练后,基于三个生物医学VQA数据集[27]对LLaVA-Med进行微调,这些数据集涵盖不同规模数据和多样化的生物医学主题。给定生物医学图像作为上下文,系统会提供多个自然语言问题,助手以自由文本形式回答闭集和开集问题,其中每个闭集问题的提示中会构建候选答案列表。
Discussion. We discuss three favorable properties/implications of LLaVA-Med: (i) Affordable development cost. Instead of scaling up data/model for the best performance, we aim to provide affordable and reasonable solutions with low development cost: it takes 7 and 8 hours for stage 1 and 2 on 8 40G A100 GPUs, respectively (see Table 5 for detailed numbers). $(i i)A$ recipe for many domains. Though this paper focuses on biomedical domains, the proposed adaptation procedure is general iz able to other vertical domains such as gaming and education, where novel concepts and domain knowledge are needed to build a helpful assistant. Similar to the don’t stop pre-training argument in [11], we consider a scalable pipeline to create domain-specific instruct data from large unlabelled data, and advocate don’t stop instruction-tuning to build customized LMM. (iii) Low serving cost. While the model size of general LMM can be giant and serving cost can be prohibitively high, customized LMM has its unique advantages in low serving cost. (iv) Smooth Model Adaptation. Alternatively, the network architecture allows us to initialize the vision encoder from BioMedCLIP [49], or initialize the language model from Vicuna [43], which may lead to higher performance. However, adapting from LLaVA smooth adaptation as a chatbot, where model’s behaviors transit from layperson to a professional assistant that is able to provide helpful domain-specific response.
讨论。我们探讨LLaVA-Med的三个优势特性/影响:(i) 可负担的开发成本。我们不追求通过扩大数据/模型规模来获得最佳性能,而是旨在提供开发成本低廉的合理解决方案:在8块40G A100 GPU上,阶段1和阶段2分别耗时7小时和8小时(具体数据见表5)。$(ii)A$ 多领域适配方案。虽然本文聚焦生物医学领域,但提出的适配流程可推广至游戏、教育等需要新概念和领域知识来构建实用助手的垂直领域。类似文献[11]提出的"不要停止预训练"观点,我们设计了一个可扩展的流程来从大规模无标注数据生成领域特定的指令数据,并主张通过"不要停止指令微调"来构建定制化大型多模态模型。(iii) 低服务成本。通用大型多模态模型的参数量可能极其庞大,导致服务成本过高,而定制化模型在低服务成本方面具有独特优势。(iv) 平滑模型适配。网络架构允许我们从BioMedCLIP[49]初始化视觉编码器,或从Vicuna[43]初始化语言模型,这可能带来更高性能。但采用LLaVA进行平滑适配时,模型行为会从外行自然过渡到能够提供专业领域响应的智能助手。
5 Experiments
5 实验
We conduct experiments to study two key components, the quality of the produced multimodal biomedical instruction-following data, and performance of LLaVA-Med. We consider two research evaluation settings: (1) What is the performance of LLaVA-Med as an open-ended biomedcal visual chatbot? (2) How does LLaVA-Med compare to existing methods on standard benchmarks? To clarify, throughout the entire experiments, we only utilize the language-only GPT-4.
我们通过实验研究两个关键组成部分:生成的多模态生物医学指令跟随数据的质量,以及LLaVA-Med的性能表现。我们考虑两种研究评估设置:(1) LLaVA-Med作为开放式生物医学视觉聊天机器人的性能如何?(2) LLaVA-Med在标准基准测试中与现有方法相比表现如何?需要说明的是,在整个实验过程中,我们仅使用了纯文本版本的GPT-4。
5.1 Biomedical Visual Chatbot
5.1 生物医学视觉聊天机器人
To evaluate the performance of LLaVA-Med on biomedical multimodal conversation, we construct an evaluation dataset with 193 novel questions. For this test dataset, we randomly selected 50 unseen image and caption pairs from PMC-15M, and generate two types of questions: conversation and detailed description. The conversation data is collected using the same self-instruct data generation pipeline as for the 2nd stage. Detailed description questions were randomly selected from a fixed set [24] of questions to elicit detailed description responses.
为了评估LLaVA-Med在生物医学多模态对话中的表现,我们构建了一个包含193个新问题的评估数据集。针对该测试集,我们从PMC-15M中随机选取了50组未见过的图像及标题对,并生成两类问题:对话型问题和详细描述型问题。对话数据采用与第二阶段相同的自指令数据生成流程进行收集。详细描述问题则从固定问题集[24]中随机选取,以获取详细描述性回答。
We leverage GPT-4 to quantify the correctness of the model answer to a question when given the image context and caption. GPT-4 makes a reference prediction, setting the upper bound answer for the teacher model. We then generate response to the same question from another LMM. Given responses from the two assistants (the candidate LMM and GPT-4), the question, figure caption, and figure context, we ask GPT-4 to score the helpfulness, relevance, accuracy, and level of details of the responses from the two assistants, and give an overall score on a scale of 1 to 10, where a higher score indicates better overall performance. GPT-4 is also asked to provide a comprehensive explanation the evaluation, for us to better understand the models. We then compute the relative score using GPT-4 reference score for normalization.
我们利用GPT-4在给定图像上下文和标题时量化模型对问题回答的正确性。GPT-4生成参考预测,为教师模型设定答案上限。接着,我们从另一个大语言模型生成对同一问题的回答。给定两个助手(候选大语言模型和GPT-4)的回答、问题、图标题和图像上下文,我们要求GPT-4对两个回答的有用性、相关性、准确性和细节程度进行评分,并按1到10分给出总体评分(分数越高表示整体表现越好)。同时要求GPT-4提供评估的详细解释,以便我们更好地理解模型性能。最后,我们使用GPT-4的参考分数进行归一化计算相对得分。
| QuestionTypes | Domains | Overall | ||||||
| Conversation | Description | CXR | MRI | Histology | Gross | CT | ||
| (Question Count) LLaVA | (143) 39.4 | (50) 26.2 | (37) 41.6 | (38) 33.4 | (44) 38.4 | (34) 32.9 | (40) 33.4 | (193) 36.1 |
| LLaVA-Med | ||||||||
| Stage 1 | 22.6 | 25.2 | 25.8 | 19.0 | 24.8 | 24.7 | 22.2 | 23.3 |
| 10K | 42.4 | 32.5 | 46.1 | 36.7 | 43.5 | 34.7 | 37.5 | 39.9 |
| 60K | 53.7 | 36.9 | 57.3 | 39.8 | 49.8 | 47.4 | 52.4 | 49.4 |
| 60K-IM | 55.1 | 36.4 | 56.2 | 40.4 | 52.7 | 51.8 | 50.1 | 50.2 |
Table 1: Performance comparison of mulitmodal chat instruction-following abilities, measured by the relative score via language GPT-4 evaluation.
| 问题类型 | 领域 | 总体 | ||||||
|---|---|---|---|---|---|---|---|---|
| 对话 | 描述 | CXR | MRI | 组织学 | 大体 | CT | ||
| (问题数量) LLaVA | (143) 39.4 | (50) 26.2 | (37) 41.6 | (38) 33.4 | (44) 38.4 | (34) 32.9 | (40) 33.4 | (193) 36.1 |
| LLaVA-Med | ||||||||
| 阶段1 | 22.6 | 25.2 | 25.8 | 19.0 | 24.8 | 24.7 | 22.2 | 23.3 |
| 10K | 42.4 | 32.5 | 46.1 | 36.7 | 43.5 | 34.7 | 37.5 | 39.9 |
| 60K | 53.7 | 36.9 | 57.3 | 39.8 | 49.8 | 47.4 | 52.4 | 49.4 |
| 60K-IM | 55.1 | 36.4 | 56.2 | 40.4 | 52.7 | 51.8 | 50.1 | 50.2 |
表 1: 多模态聊天指令跟随能力的性能比较,通过语言GPT-4评估的相对得分衡量。
Visual input example, Biomedical image from [6]
视觉输入示例,生物医学图像来自 [6]
| Caption:Chest x-ray(CXR) on Day 2 of | |
| User LLaVA-Med User LLaVA-Med procedures. | seenthroughoutthelungswithrelative subpleuralsparing(redarrows) Source: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7398722/ Whatisshowninthisimage? The image is a chest X-ray (CXR) that shows bilateral patchy infiltrates,which are areas of increased opacity in the lungs. These infiltrates can be indicative of various lung conditions, such as infections, inflammation,or otherlung diseases. Are there any devices implanted in this patient? Yes, it appears that the patient has several devices implanted. These devices may include endotracheal tubes,central venous catheters, and/or pulmonary artery catheters.These devices are typically used for monitoring and managing various medical conditions, as well as for providing support during surgical What is shown in this image? |
| User LLaVA | |
| diagram. User LLaVA for treatment or monitoring. | The image shows a ribcage with wires coming out of it, indicating that it is likely a medical illustration or Are there any devices implanted in this patient? Yes, there are several devices protruding from the patient's chest, which are likely medical devices used |
| User Language GPT-4 | What is shown in this image? Theimageshows a chest x-ray taken onDay 2of apatient's admission after intubation.You cansee the endotracheal tube marked by a yellow line, and rapidly worsening ground-glass opacities throughout the lungs. There is also relative subpleural sparing, which is indicated by red arrows. |
Table 2: Example comparison of medical visual chat and reasoning capabilities. The language-only GPT-4 is considered as the performance upper bound, as the golden captions and inline mentions are fed into GPT-4 as the context, without requiring the model to understand the raw image.
表 2: 医疗视觉对话与推理能力示例对比。纯语言模型 GPT-4 被视为性能上限,因为黄金标注和文中提及内容会作为上下文输入 GPT-4,无需模型理解原始图像。
The results are reported in Table 1. LLaVA-Med with Stage-1 training alone is insufficient as a chatbot, as it loses its ability to follow diverse instructions, though biomedical concept coverage is improved. LLaVA-Med with the full two-stage training consistently outperforms the general domain LLaVA, and training with larger instruct data (from 10K to 60K samples) leads to higher performance. When inline mentions are considered in self-instruct, the generated data 60K-IM slightly improves the chat ability. The results demonstrate the effectiveness of the strategies in biomedical instructionfollowing data collection as well as the value of dataset assets. Overall, for the best LLaVA-Med, it matches the $50.2%$ performance of GPT-4. Note that GPT-4 generates response by considering ground-truth caption and golden inline mentions, without understanding the images. Though not a fair comparison between LMMs and GPT-4, GPT-4 is a consistent and reliable evaluation tool.
结果如表 1 所示。仅进行第一阶段训练的 LLaVA-Med 作为聊天机器人表现不足,虽然生物医学概念覆盖有所提升,但丧失了遵循多样化指令的能力。完整两阶段训练的 LLaVA-Med 始终优于通用领域的 LLaVA,且使用更大规模指令数据 (从 10K 到 60K 样本) 训练会带来更高性能。当自指令生成考虑内联提及时,60K-IM 生成数据能略微提升对话能力。这些结果证明了生物医学指令跟随数据收集策略的有效性以及数据集资产的价值。最佳性能的 LLaVA-Med 达到了 GPT-4 50.2% 的水平。需注意 GPT-4 在生成响应时会参考真实标注和黄金内联提及,但并不理解图像内容。尽管大语言模型与 GPT-4 的对比并不公平,但 GPT-4 仍是稳定可靠的评估工具。
In Table 2, we provide examples on the biomed visual conversations of different chatbots. LLaVAMed precisely answers the questions with biomedical knowledge, while LLaVA behaves like a layperson, who hallucinate based on commonsense. Since the multimodal GPT-4 is not publicly available, we resort to language-only GPT-4 for comparison. We feed golden captions and inline mentions into GPT-4 as the context, it generates knowledgeable response through re-organizing the information in the conversational manner.
在表2中,我们提供了不同聊天机器人在生物医学视觉对话中的示例。LLaVAMed能基于生物医学知识准确回答问题,而LLaVA表现得像外行,会基于常识产生幻觉。由于多模态GPT-4尚未公开,我们采用纯语言版本的GPT-4进行对比。将黄金标注和文中提及内容作为上下文输入GPT-4后,它能通过对话式重组信息生成专业回答。
5.2 Performance on Established Benchmarks
5.2 在现有基准测试上的表现
Dataset Description. We train and evaluate LLaVA-Med on three biomedical VQA datasets. The detailed data statistics are summarized in Table 3.
数据集描述。我们在三个生物医学视觉问答(VQA)数据集上训练和评估LLaVA-Med。详细数据统计信息总结如表3所示。
• VQA-RAD [16] contains $3515\mathrm{QA}$ pairs generated by clinicians and 315 radiology images that are evenly distributed over the head, chest, and abdomen. Each image is associated with multiple questions. Questions are categorized into 11 categories: abnormality, attribute, modality, organ system, color, counting, object/condition presence, size, plane, positional reasoning, and other. Half of the answers are closed-ended (i.e., yes/no type), while the rest are open- ended with either one-word or short phrase answers.
• VQA-RAD [16] 包含由临床医生生成的3515个QA对和315张均匀分布在头部、胸部和腹部的放射影像。每张影像关联多个问题,问题分为11类:异常 (abnormality)、属性 (attribute)、模态 (modality)、器官系统 (organ system)、颜色 (color)、计数 (counting)、对象/条件存在 (object/condition presence)、尺寸 (size)、平面 (plane)、位置推理 (positional reasoning) 及其他 (other)。半数答案为封闭式(即是非型),其余为开放式(需回答单词或短语)。
| Dataset | VQA-RAD Train Test | SLAKE | PathVQA | |||||
| Train | Val | Test | Train | Val | Test | |||
| # Images | 313 | 203 | 450 | 96 | 96 | 2599 | 858 | 858 |
| #QAPairs | 1797 | 451 | 4919 | 1053 | 1061 | 19,755 | 6279 | 6761 |
| #Open | 770 | 179 | 2976 | 631 | 645 | 9949 | 3144 | 3370 |
| #Closed | 1027 | 272 | 1943 | 422 | 416 | 9806 | 3135 | 3391 |
Table 3: Dataset statistics. For SLAKE, only the English subset is considered for head-to-head comparison with existing methods.
| Dataset | VQA-RAD Train Test | SLAKE | PathVQA | |||||
|---|---|---|---|---|---|---|---|---|
| Train | Val | Test | Train | Val | Test | |||
| # Images | 313 | 203 | 450 | 96 | 96 | 2599 | 858 | 858 |
| #QAPairs | 1797 | 451 | 4919 | 1053 | 1061 | 19,755 | 6279 | 6761 |
| #Open | 770 | 179 | 2976 | 631 | 645 | 9949 | 3144 | 3370 |
| #Closed | 1027 | 272 | 1943 | 422 | 416 | 9806 | 3135 | 3391 |
表 3: 数据集统计。对于SLAKE数据集,仅考虑英文子集以便与现有方法进行直接对比。
• SLAKE [23] is a Semantically-Labeled Knowledge-Enhanced dataset for medical VQA. It consists of 642 radiology images and over 7000 diverse QA pairs annotated by experienced physicians, where the questions may involve external medical knowledge (solved by provided medical knowledge graph), and the images are associated with rich visual annotations, including semantic segmentation masks and object detection bounding boxes. Besides, SLAKE includes richer modalities and covers more human body parts than the currently available dataset, including brain, neck, chest, abdomen, and pelvic cavity. Note SLAKE is bilingual dataset with English and Chinese. When compared with existing methods, we only consider the English subset. • PathVQA [13] is a dataset of pathology images. It c
