Post-translational modification prediction via prompt-based fine-tuning of a GPT-2 model
基于GPT-2模型的提示微调实现翻译后修饰预测
Received: 11 March 2024
收到日期:2024年3月11日
Accepted: 29 July 2024
录用日期: 2024年7月29日
Published online: 07 August 2024
在线发布:2024年8月7日
Check for updates
检查更新
Palistha Shrestha 1,5, Jeevan Kandel 2,5, Hilal Tayara 3 & Kil To Chong 1,4
Palistha Shrestha 1,5, Jeevan Kandel 2,5, Hilal Tayara 3 & Kil To Chong 1,4
Post-translational modifications (PTMs) are pivotal in modulating protein functions and influencing cellular processes like signaling, localization, and degradation. The complexity of these biological interactions necessitates efficient predictive methodologies. In this work, we introduce PTMGPT2, an interpret able protein language model that utilizes prompt-based fine-tuning to improve its accuracy in precisely predicting PTMs. Drawing inspiration from recent advancements in GPT-based architectures, PTMGPT2 adopts unsupervised learning to identify PTMs. It utilizes a custom prompt to guide the model through the subtle linguistic patterns encoded in amino acid sequences, generating tokens indicative of PTM sites. To provide interpret ability, we visualize attention profiles from the model’s final decoder layer to elucidate sequence motifs essential for molecular recognition and analyze the effects of mutations at or near PTM sites to offer deeper insights into protein functionality. Comparative assessments reveal that PTMGPT2 outperforms existing methods across 19 PTM types, underscoring its potential in identifying disease associations and drug targets.
翻译后修饰 (PTM) 在调节蛋白质功能和影响细胞过程 (如信号传导、定位和降解) 中起着关键作用。这些生物相互作用的复杂性需要高效的预测方法。在这项工作中,我们引入了 PTMGPT2,这是一种可解释的蛋白质语言模型,它利用基于提示的微调来提高其精确预测 PTM 的准确性。受 GPT 架构最新进展的启发,PTMGPT2 采用无监督学习来识别 PTM。它利用自定义提示来引导模型理解氨基酸序列中编码的微妙语言模式,生成指示 PTM 位点的 Token。为了提供可解释性,我们可视化模型最后解码层的注意力分布,以阐明对分子识别至关重要的序列模体,并分析 PTM 位点处或附近突变的影响,以提供对蛋白质功能的更深入理解。比较评估表明,PTMGPT2 在 19 种 PTM 类型中优于现有方法,突显了其在识别疾病关联和药物靶点方面的潜力。
Proteins, the essential workhorses of the cell, are modulated by posttranslational modifications (PTM), a process vital for their optimal functioning. With over 400 known types of PTMs1, they enhance the functional spectrum of the proteome. Despite originating from a foundational set of approximately 20,000 protein-coding genes2, the dynamic realm of PTMs is estimated to expand the human proteome to over a million unique protein species. This diversity echoes the complexity inherent in human languages: individual amino acids assemble into’words’ to form functional’sentences’ or domains. This linguistic parallel extends to the dense, information-rich structure of both protein sequences and human languages. The advancing field of Natural Language Processing (NLP) not only unravels the intricacies of human communication but is also increasingly applied to decode the complex language of proteins. Our study leverages the transformative capabilities of generative pretrained transformers (GPT) based models, a cornerstone in NLP, to interpret and predict the complex landscape of PTMs, highlighting an intersection where computational linguistics meets molecular biology.
蛋白质作为细胞中不可或缺的主力军,其功能通过翻译后修饰 (PTM) 这一关键过程进行调控,以确保它们发挥最佳作用。目前已知的PTM类型超过400种1,它们拓展了蛋白质组的功能范围。尽管源自约20,000个蛋白质编码基因2这一基础集合,PTM的动态领域预计将人类蛋白质组扩展至超过百万种独特蛋白质变体。这种多样性映照了人类语言与生俱来的复杂性:单个氨基酸组装成"词汇",进而形成功能性的"句子"或结构域。这种语言学上的相似性延伸至蛋白质序列和人类语言共有的密集且信息丰富的结构特征。自然语言处理 (NLP) 领域的发展不仅揭示了人类交流的复杂性,也日益应用于解读蛋白质的复杂语言。我们的研究利用基于生成式预训练Transformer (GPT) 模型的变革性能力——这一NLP领域的基石技术,来解读和预测PTM的复杂景观,凸显了计算语言学与分子生物学交叉融合的前沿领域。
In the quest to predict PTM sites, the scientific community has predominantly relied on supervised methods, which have evolved significantly over the years3–5. These methods typically involve training algorithms on datasets where the modification status of each site is known, allowing the model to learn and predict modifications on new sequences. B.Trost and A.Kusalik6 initially focus on methods like Support Vector Machines and decision trees, which classify amino acid sequences based on geometric margins or hierarchical decision rules. Progressing towards more sophisticated approaches, Zhou, F. et al. 7 discuss the utilization of Convolutional Neural Networks and
在预测翻译后修饰 (PTM) 位点的研究中,科学界主要依赖于监督学习方法,这些方法经过多年发展已取得显著进步[3-5]。这类方法通常基于已知位点修饰状态的数据集训练算法,使模型能够学习并预测新序列的修饰情况。B.Trost 和 A.Kusalik[6] 最初聚焦于支持向量机和决策树等方法,这些方法通过几何间隔或分层决策规则对氨基酸序列进行分类。随着方法日益复杂,Zhou, F. 等人[7] 探讨了卷积神经网络的应用
Recurrent Neural Networks, adept at recognizing complex patterns and capturing temporal sequence dynamics. DeepSucc8 proposed a specific deep learning architecture for identifying succ in yl ation sites, indicative of the tailored application of deep learning in PTM prediction. Smith, L. M. and Kelleher, N. L9. highlight the challenges in data representation and quality in proteomics. Building upon these developments, Smith, D. et al. 10 further advance the field, presenting a deep learning-based approach that achieves high accuracy in PTM site prediction. Their method involves intricate neural network architectures optimized for analyzing protein sequences, signifying a refined integration of deep learning in protein sequence analysis.
循环神经网络 (Recurrent Neural Networks) ,擅长识别复杂模式并捕捉时间序列动态。DeepSucc8提出了一种特定的深度学习架构,用于识别琥珀酰化 (succinylation) 位点,这体现了深度学习在翻译后修饰预测中的定制化应用。Smith, L. M. 和 Kelleher, N. L [9] 强调了蛋白质组学中数据表示和质量的挑战。基于这些进展,Smith, D. 等人 [10] 进一步推动了该领域发展,提出了一种基于深度学习的方法,在翻译后修饰位点预测中实现了高精度。他们的方法涉及复杂的神经网络架构,这些架构经过优化以分析蛋白质序列,标志着深度学习在蛋白质序列分析中的精细化整合。
On the unsupervised learning front, methods like those developed by Chung, C. et al.11 have significantly advanced the field. Central to their algorithm is the clustering of similar features and the recognition of patterns indicative of PTM sites. Lee, Tzong-Yi, et al. 12 utilized distant sequence features in combination with Radial Basis Function Networks, an approach that effectively identifies ubiquitin conjugation sites by integrating non-local sequence information. However, the complexity of PTM processes, often characterized by subtle and context-dependent patterns, pose challenges to these methods. Despite their advancements, they often grapple with issues like data imbalance, where certain PTMs are underrepresented, and the dependency on high-quality annotated datasets. These approaches can be challenged by the intricate and subtle nature of PTM sites, potentially overlooking crucial biological details. This landscape of PTM site prediction is ripe for innovation through generative transformer models, particularly in the domain of unsupervised learning. Intrigued by this possibility, we explored the potential of generative transformers, exemplified by the GPT architecture, for predicting PTM sites.
在无监督学习方面,Chung, C.等人[11]开发的方法显著推动了该领域的发展。其算法的核心在于对相似特征进行聚类,并识别指示PTM位点的模式。Lee, Tzong-Yi等人[12]利用远距离序列特征结合径向基函数网络,通过整合非局部序列信息有效识别泛素结合位点。然而PTM过程通常具有细微且依赖上下文模式的特点,这种复杂性对这些方法构成了挑战。尽管取得了进展,这些方法仍需应对数据不平衡(某些PTM代表不足)和对高质量标注数据集的依赖等问题。PTM位点错综复杂的特性可能使这些方法难以捕捉关键的生物学细节。当前PTM位点预测领域正处于通过生成式Transformer模型实现突破的成熟阶段,特别是在无监督学习领域。受此前景吸引,我们探索了以GPT架构为代表的生成式Transformer在预测PTM位点方面的潜力。
Here, we introduce PTMGPT2, a suite of models capable of generating tokens that signify modified protein sequences, crucial for identifying PTM sites. At the core of this platform is PROTGPT213, an auto regressive transformer model. We have adapted PROTGPT2, utilizing it as a pre-trained model, and further fine-tuned it for the specific task of generating classification labels for a given PTM type. PTMGPT2 utilizes a decoder-only architecture, which eliminates the need for a task-specific classification head during training. Instead, the final layer of the decoder functions as a projection back to the vocabulary space, effectively generating the next possible token based on the learned patterns among tokens in the input prompt. When provided with a prompt, the model is faced with a protein sequence structured in a fillin-the-blank format. Impressively, even without any hyper parameter optimization procedures, our model has demonstrated an average $5.45%$ improvement in Matthews Correlation Coefficient (MCC) over all other competing methods. The webserver and models that underpin PTMGPT2 are available at https://nsclbio.jbnu.ac.kr/tools/ ptmgpt2. Given the critical role of PTM in elucidating the mechanisms of various biological processes, we believe PTMGPT2 represents a significant stride forward in the efficient prediction and analysis of protein sequences.
在此,我们推出PTMGPT2,这是一套能够生成表示修饰蛋白质序列的token的模型,对于识别PTM位点至关重要。该平台的核心是PROTGPT213,一种自回归Transformer模型。我们调整了PROTGPT2,将其用作预训练模型,并进一步针对生成特定PTM类型分类标签的任务进行了微调。PTMGPT2采用仅解码器架构,在训练过程中无需特定任务的分类头。相反,解码器的最后一层作为投影层返回词汇空间,基于输入提示中token之间的学习模式,有效地生成下一个可能的token。当提供提示时,模型面对的是以填空格式构建的蛋白质序列。令人印象深刻的是,即使没有任何超参数优化过程,我们的模型在马修斯相关系数 (MCC) 上比所有其他竞争方法平均提高了 $5.45%$ 。支持PTMGPT2的网站服务器和模型可在 https://nsclbio.jbnu.ac.kr/tools/ ptmgpt2 获取。鉴于PTM在阐明各种生物过程机制中的关键作用,我们相信PTMGPT2代表了在蛋白质序列高效预测和分析方面的重要进展。
Results
结果
PTMGPT2 implements a prompt-based approach for PTM prediction
PTMGPT2 采用基于提示的方法进行翻译后修饰 (PTM) 预测
We introduce an end-to-end deep learning framework depicted in Fig. 1, utilizing a GPT as the foundational model. Central to our approach is the prompt-based finetuning of the PROTGPT2 model in an unsupervised manner. This is achieved by utilizing informative prompts during training, enabling the model to generate accurate sequence labels. The design of these prompts is a critical aspect of our architecture, as they provide essential instructional input to the pretrained model, guiding its learning process. To enhance the explanatory power of these prompts, we have introduced four custom tokens to the pre-trained tokenizer, expanding its vocabulary size from 50,257 to 50,264. This modification is particularly significant due to the tokenizer’s reliance on the Byte Pair Encoding (BPE) algorithm 14. A notable consequence of this approach is that our model goes beyond annotating individual amino acid residues. Instead, it focuses on annotating variable-length protein sequence motifs. This strategy is pivotal as it ensures the preservation of evolutionary biological function ali ties, allowing for a more nuanced and biologically relevant interpretation of protein sequences.
我们引入了一个端到端的深度学习框架 (如图 1 所示) , 使用 GPT 作为基础模型。我们方法的核心是以无监督方式对 PROTGPT2 模型进行基于提示的微调。这是通过在训练期间使用信息性提示来实现的, 使模型能够生成准确的序列标签。这些提示的设计是我们架构的一个关键方面, 因为它们为预训练模型提供了必要的指令输入, 指导其学习过程。为了增强这些提示的解释能力, 我们向预训练的分词器引入了四个自定义 token, 将其词汇表大小从 50,257 扩展到 50,264。由于该分词器依赖于字节对编码 (BPE) 算法 [14], 这一修改尤为重要。这种方法的一个显著结果是, 我们的模型不仅能够注释单个氨基酸残基, 还能专注于注释可变长度的蛋白质序列 motif。该策略至关重要, 因为它确保了进化生物学功能特性的保留, 从而实现对蛋白质序列更细致且更具生物学意义的解读。
In the PTMGPT2 framework, we employ a prompt structure that incorporates four principal tokens. The first, designated as the ‘SEQUENCE:’ token, represents the specific protein sub sequence of interest. The second, known as the ‘LABEL:’ token, indicates whether the sub sequence is modified (‘POSITIVE’) or unmodified (‘NEGATIVE’). This token-driven prompt design forms the foundation for the fine-tuning process of the PTMGPT2 model, enabling it to accurately generate labels during inference. A key aspect of this model lies in its architectural foundation, which is based on GPT $\cdot2^{15}$ . This architecture is characterized by its exclusive use of decoder layers, with PTMGPT2 utilizing a total of 36 such layers, consistent with the pretrained model. This maintains architectural consistency while fine-tuning for our downstream task of PTM site prediction. Each of these layers is composed of masked self-attention mechanisms 16, which ensure that during the training phase, the protein sequence and custom tokens can be influenced only by their preceding tokens in the prompt. This is essential for maintaining the auto regressive property of the model. Such a method is fundamental for our model’s ability to accurately generate labels, as it helps preserve the chronological integrity of biological sequence data and its dependencies with custom tokens, ensuring that the predictions are biologically relevant.
在PTMGPT2框架中, 我们采用包含四个主要token的提示结构。第一个是 "SEQUENCE:" token, 代表目标蛋白质亚序列。第二个是 "LABEL:" token, 指示该亚序列是否被修饰 ("POSITIVE") 或未被修饰 ("NEGATIVE") 。这种基于token的提示设计构成了PTMGPT2模型微调过程的基础, 使其能够在推理过程中准确生成标签。该模型的一个关键特性在于其架构基础基于GPT $\cdot2^{15}$ 。该架构的特点是仅使用解码器层, PTMGPT2共使用36个这样的层, 与预训练模型保持一致。这在对我们的PTM位点预测下游任务进行微调时保持了架构一致性。每个层都由掩码自注意力机制16组成, 确保在训练阶段, 蛋白质序列和自定义token仅受提示中前序token的影响。这对于保持模型的自回归特性至关重要。这种方法对我们模型准确生成标签的能力至关重要, 因为它有助于保持生物序列数据的时间完整性及其与自定义token的依赖关系, 确保预测结果具有生物学意义。
A key distinction in our approach lies in the methodology we employed for prompt based fine-tuning during the training and inference phases of PTMGPT2. During the training phase, PTMGPT2 is engaged in an unsupervised learning process. This approach involves feeding the model with input prompts and training it to output the same prompt, thereby facilitating the learning of token relationships and context within the prompts themselves. This process enables the model to generate the next token based on the patterns learned during training between protein sub sequences and their corresponding labels. The approach shifts during the inference phase, where the prompts are modified by removing the ‘POSITIVE’ and ‘NEGATIVE’ tokens, effectively turning these prompts into a fill-in-the-blank exercise for the model. This strategic masking triggers PTMGPT2 to generate the labels independently, based on the patterns and associations it learned during the training phase. An essential aspect of our prompt structure is the consistent inclusion of the ‘<start of text $>^{\prime}$ and ‘<endoftext $>^{\prime}$ tokens. These tokens are integral to our prompts, signifying the beginning and end of the prompt helping the model to contextual ize the input more effectively. This interplay of training techniques and strategic prompt structuring enables PTMGPT2 to achieve high prediction accuracy and efficiency. Such an approach sets PTMGPT2 apart as an advanced tool for protein sequence analysis, particularly in predicting PTMs.
我们方法的一个关键区别在于PTMGPT2训练和推理阶段采用的基于提示的微调方法。在训练阶段,PTMGPT2进行无监督学习,通过输入提示训练模型输出相同提示,从而学习token关系及提示内部的上下文关联。这使得模型能够根据训练中学到的蛋白质子序列与其对应标签之间的模式来生成下一个token。在推理阶段,该方法发生转变:通过移除"POSITIVE"和"NEGATIVE"token将提示转化为填空任务,这种策略性掩码触发PTMGPT2基于训练阶段学习的模式关联独立生成标签。我们提示结构的重要特点是始终包含<start of text \$>^{\prime}\$和<endoftext \$>^{\prime}\$token,这些标记提示的起止位置,帮助模型更有效地理解输入上下文。这种训练技术与策略性提示结构的结合,使PTMGPT2实现了高预测精度和效率,使其成为蛋白质序列分析(尤其是翻译后修饰预测)的先进工具。
Effect of prompt design and fine-tuning on PTMGPT2 performance
提示设计和微调对PTMGPT2性能的影响
We designed five prompts with custom tokens (‘SEQUENCE:’, ‘LABEL:’, ‘POSITIVE’, and ‘NEGATIVE’) to identify the most efficient one for capturing complexity, allowing PTMGPT2 to learn and process specific sequence segments for more meaningful representations. Initially, we crafted a prompt that integrates all custom tokens with a 21-length protein sub sequence. Subsequent explorations were conducted with 51-length sub sequence and 21-length sub sequence split into groups of k-mers, with and without the custom tokens. Considering that the pretrained model was originally trained solely on protein sequences, we
我们设计了五个包含自定义标记 ( 'SEQUENCE:', 'LABEL:', 'POSITIVE', 'NEGATIVE' ) 的提示方案, 以确定最能有效捕捉复杂性的方案, 使 PTMGPT2 能够学习并处理特定序列片段, 从而获得更有意义的表征。最初, 我们构建了一个整合所有自定义标记与 21 长度蛋白质子序列的提示方案。后续探索采用 51 长度子序列, 以及将 21 长度子序列拆分为 k-mer 组的形式, 分别测试了包含与不包含自定义标记的情况。考虑到预训练模型最初仅基于蛋白质序列进行训练, 我们
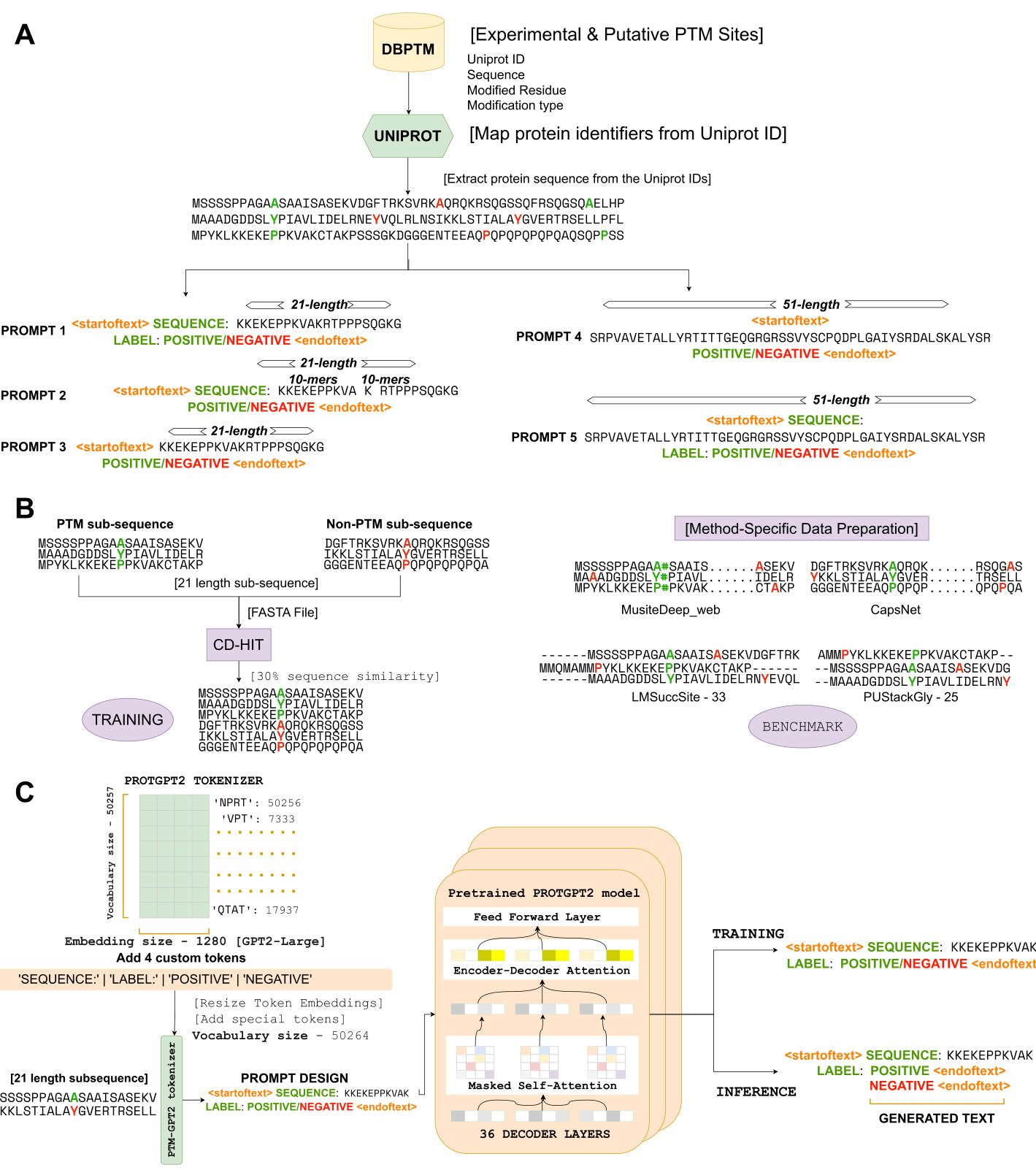
Fig. 1 | Schematic representation of the PTMGPT2 framework. A Preparation of inputs for PTMGPT2, detailing the extraction of protein sequences from Uniprot and the generation of five distinct prompt designs. B Method-specific data preparation process for benchmark, depicting both modified and unmodified subsequence extraction, followed by the creation of a training dataset using CD-HIT for

图 1 | PTMGPT2 框架示意图。A PTMGPT2 的输入数据准备流程,详细说明从 Uniprot 提取蛋白质序列及生成五种不同提示设计的过程。B 基准测试的特定方法数据准备流程,展示修饰与未修饰子序列的提取步骤,随后通过 CD-HIT 构建训练数据集。
$30%$ sequence similarity C Architecture of the PTMGPT2 model and the training and inference processes. It highlights the integration of custom tokens into the tokenizer, the resizing of token embeddings, and the subsequent prompt design utilized during training and inference to generate predictions.
$30%$ 序列相似性 C PTMGPT2 模型架构及训练与推理流程。该图展示了将自定义 token 集成至分词器、调整 token 嵌入维度,以及在训练和推理过程中用于生成预测的后续提示设计。
fine-tuned it with prompts both with and without the tokens to ascertain their actual contribution to improving PTM predictions.
通过使用包含和不包含这些token的提示进行微调, 以确定它们对改进PTM预测的实际贡献。
Upon fine-tuning PTMGPT2 with training datasets for arginine(R) methyl ation and tyrosine(Y) phosphor yl ation, it became evident that the prompt containing the 21-length sub sequence and the four custom tokens yielded the best results in generating accurate labels, as shown in Table 1. For methyl ation (R), the MCC, F1 Score, precision, and recall were reported as 80.51, 81.32, 95.14, and 71.01, respectively. Similarly, for phosphor yl ation (Y), the MCC, F1 Score, precision, and recall were
在使用精氨酸 (R) 甲基化和酪氨酸 (Y) 磷酸化训练数据集对 PTMGPT2 进行微调后,我们发现包含 21 长度子序列和四个自定义 token 的提示在生成准确标签方面效果最佳,如表 1 所示。对于甲基化 (R) ,其 MCC、F1 分数、精确度和召回率分别为 80.51、81.32、95.14 和 71.01。同样地,对于磷酸化 (Y) ,其 MCC、F1 分数、精确度和召回率分别为
48.83, 46.98, 30.95, and 97.51, respectively. So, for all the experiments, we used the 21-length sequence with custom tokens. The inclusion of ‘SEQUENCE:’ and ‘LABEL:’ tokens provided clear contextual cues to the model, allowing it to understand the structure of the input and the expected output format. This helped the model differentiate between the sequence data and the classification labels, leading to better learning and prediction accuracy. The 21-length sub sequence was an ideal size for the model to capture the necessary information without being too short to miss important context or too long to introduce noise. By framing the task clearly with the ‘SEQUENCE:’ and ‘LABEL:’ tokens, the model faced less ambiguity in generating predictions, which can be particularly beneficial for complex tasks such as PTM site prediction.
分别为48.83、46.98、30.95和97.51。因此在所有实验中, 我们采用了包含自定义token的21长度序列。"SEQUENCE:"和"LABEL:" token的加入为模型提供了清晰的上下文线索, 使其能够理解输入数据的结构和预期输出格式。这有助于模型区分序列数据和分类标签, 从而提升学习效果和预测准确性。21长度子序列是理想尺寸, 既能确保模型捕获必要信息, 又避免了因过短而遗漏关键上下文或因过长而引入噪声。通过"SEQUENCE:"和"LABEL:" token明确界定任务, 模型在生成预测时面临的模糊性显著降低, 这对于像PTM位点预测这类复杂任务尤为有利。
Table 1 | Benchmark results of PTMGPT2 after fine-tuning for optimal prompt selection
| PTM | Prompt | MCC F1Score | Precision | Recall |
| Methylation(R) | 21-lengthw/tokens[Proposed] 80.51 | 81.32 | 95.14 | 71.01 |
| 21-lengthk-merw/otokens 77.16 | 78.53 | 94.98 | 65.03 | |
| 51-lengthw/otokens | 58.25 63.37 | 85.02 | 33.56 | |
| 51-lengthw/tokens | 60.85 66.98 | 89.34 | 45.77 | |
| Phosphorylation(Y) | 21-lengthw/tokens[Proposed] | 48.83 46.98 | 30.95 | 97.51 |
| 21-lengthw/otokens | 45.27 44.07 | 30.04 | 90.47 | |
| 51-lengthw/otokens | 27.48 31.25 | 20.32 | 67.56 | |
| 51-lengthw/tokens | 31.01 32.76 | 20.71 | 78.37 |
Top values are represented in bold.
表 1 | 微调后 PTMGPT2 最优提示选择的基准测试结果
| PTM | Prompt | MCC F1Score | Precision | Recall |
|---|---|---|---|---|
| Methylation (R) | 21-lengthw/tokens [Proposed] | 80.51 | 81.32 | 95.14 |
| 21-lengthk-merw/otokens | 77.16 | 78.53 | 94.98 | |
| 51-lengthw/otokens | 58.25 | 63.37 | 85.02 | |
| 51-lengthw/tokens | 60.85 | 66.98 | 89.34 | |
| Phosphorylation (Y) | 21-lengthw/tokens [Proposed] | 48.83 | 46.98 | 30.95 |
| 21-lengthw/otokens | 45.27 | 44.07 | 30.04 | |
| 51-lengthw/otokens | 27.48 | 31.25 | 20.32 | |
| 51-lengthw/tokens | 31.01 | 32.76 | 20.71 |
最高值以粗体表示。
Comparative benchmark analysis reveals PTMGPT2’s dominance
比较基准分析显示 PTMGPT2 占据主导地位
To validate PTMGPT2’s performance, benchmarking against a database that encompasses a broad spectrum of experimentally verified PTMs and annotates potential PTMs for all UniProt17 entries was imperative. Accordingly, we chose the DBPTM database18 for its extensive collection of benchmark datasets, tailored for distinct types of PTMs. The inclusion of highly imbalanced datasets from DBPTM proved to be particularly advantageous, as it enabled a precise evaluation of PTMGPT2s ability to identify unmodified amino acid residues. This capability is crucial, considering that the majority of residues in a protein sequence typically remain unmodified. For a thorough assessment, we sourced 19 distinct benchmarking datasets from DBPTM, each containing a minimum of 500 data points corresponding to a specific PTM type.
为验证 PTMGPT2 的性能,必须使用包含广泛实验验证的翻译后修饰 (PTM) 并注释所有 UniProt17 条目潜在 PTM 的数据库进行基准测试。因此,我们选择 DBPTM 数据库 [18] ,其专为不同类型的 PTM 定制了广泛的基准数据集。DBPTM 中高度不平衡数据集的纳入被证明特别有利,因为它能精确评估 PTMGPT2 识别未修饰氨基酸残基的能力。考虑到蛋白质序列中大多数残基通常保持未修饰状态,这一能力至关重要。为进行全面评估,我们从 DBPTM 获取了 19 个不同的基准数据集,每个数据集至少包含 500 个对应特定 PTM 类型的数据点。
Our comparative analysis underscores PTMGPT2’s capability in predicting a variety of PTMs, marking substantial improvements when benchmarked against established methodologies using the MCC as the metric as shown in Table 2. For instance, in the case of lysine(K) succinylation, Succ-PTMGPT2 achieved a notable $7.94%$ improvement over LM-SuccSite. In the case of lysine(K) sumo yl ation, SumoyPTMGPT2 surpassed GPS Sumo by $5.91%$ . The trend continued with N-linked g lycos yl ation on asparagine(N), where N-linked-PTMGPT2 outperformed Musite-Web by $5.62%$ . RMethyl-PTMGPT2, targeting arginine(R) methyl ation, surpassed Musite-Web by $12.74%$ . Even in scenarios with marginal gains, such as lysine(K) acetyl ation where KAcetyl-PTMGPT2 edged out Musite-web by $0.46%$ , PTMGPT2 maintained its lead. PTMGPT2 exhibited robust performance for lysine(K) ubiquitin ation, surpassing Musite-Web by $5.01%$ . It achieved a $9.08%$ higher accuracy in predicting O-linked g lycos yl ation on serine(S) and threonine(T) residues. For cysteine(C) S-nitro syl ation, the model outperformed PresSNO by $4.09%$ . In lysine(K) mal on yl ation, PTMGPT2’s accuracy exceeded that of DL-Malosite by $3.25%$ , and for lysine(K) methyl ation, it achieved $2.47%$ higher accuracy than MethylSite. Although PhosphoST-PTMGPT2’s performance in serine-threonine (S, T) phosphor yl ation prediction was $16.37%$ , lower than Musite-Web, it excelled in tyrosine(Y) phosphor yl ation with an accuracy of $48.83%$ , which was notably higher than Musite-Web’s $40.83%$ and Capsnet’s $43.85%$ . In the case of cysteine (C) glut at hi on yl ation and lysine (K) glut ary la tion, Glut at hi oPT MG PT 2 and Glutary-PTMGPT2 exhibited improvements of $7.51%$ and $6.48%$ over DeepGSH and ProtTrans-Glutar, respectively. In the case of valine (V) amidation and cysteine (C) spalm i to yl ation, Ami-PTMGPT2 and Palm-PTMGPT2 surpassed prAS and CapsNet by $4.78%$ and $1.56%$ , respectively. Similarly, in the cases of proline (P) hydroxyl ation, lysine (K) hydroxyl ation, and lysine (K)
我们的比较分析凸显了PTMGPT2在预测多种翻译后修饰方面的能力, 如表2所示, 以MCC作为评估指标时, 相较于已有方法取得了显著提升。例如在赖氨酸(K)琥珀酰化预测中, Succ-PTMGPT2较LM-SuccSite实现了7.94%的显著提升。在赖氨酸(K)苏素化预测中, SumoyPTMGPT2以5.91%的优势超越GPS-Sumo。这一优势在天冬酰胺(N)的N-连接糖基化预测中得以延续, N-linked-PTMGPT2以5.62%的准确率超越Musite-Web。针对精氨酸(R)甲基化的RMethyl-PTMGPT2则以12.74%的幅度领先Musite-Web。即使在提升幅度较小的场景中, 如赖氨酸(K)乙酰化预测中KAcetyl-PTMGPT2仅以0.46%的微弱优势超越Musite-Web, PTMGPT2仍保持领先地位。
PTMGPT2在赖氨酸(K)泛素化预测中展现出稳健性能, 以5.01%的优势超越Musite-Web。在丝氨酸(S)和苏氨酸(T)残基的O-连接糖基化预测中, 其准确率提升达9.08%。对于半胱氨酸(C)S-亚硝基化预测, 该模型以4.09%的优势超越PresSNO。在赖氨酸(K)丙二酰化预测中, PTMGPT2的准确率较DL-Malosite提升3.25%, 在赖氨酸(K)甲基化预测中较MethylSite提升2.47%。尽管PhosphoST-PTMGPT2在丝氨酸-苏氨酸(S,T)磷酸化预测中的表现较Musite-Web低16.37%, 但在酪氨酸(Y)磷酸化预测中以48.83%的准确率显著优于Musite-Web的40.83%和Capsnet的43.85%。
在半胱氨酸(C)谷胱甘肽化和赖氨酸(K)戊二酰化预测中, GlutathioPTMGPT2和Glutary-PTMGPT2分别较DeepGSH和ProtTrans-Glutar提升7.51%和6.48%。在缬氨酸(V)酰胺化和半胱氨酸(C)棕榈酰化预测中, Ami-PTMGPT2和Palm-PTMGPT2分别以4.78%和1.56%的优势超越prAS和CapsNet。类似地, 在脯氨酸(P)羟基化、赖氨酸(K)羟基化等预测任务中...
for myl ation, PTMGPT2 achieved superior performance over CapsNet by $11.02%$ , $7.58%$ , and $4.39%$ , respectively. Collectively, these results demonstrate the significant progress made by PTMGPT2 in advancing the precision of PTM site prediction, thereby solidifying its place as a leading tool in proteomics research.
在我的评估中,PTMGPT2相较于CapsNet分别实现了11.02%、7.58%和4.39%的性能提升。这些结果共同证明了PTMGPT2在推进PTM位点预测精度方面取得的显著进展,从而巩固了其在蛋白质组学研究中的领先工具地位。
PTMGPT2 captures sequence-label dependencies through an attention-driven interpret able framework
PTMGPT2通过注意力驱动的可解释框架捕捉序列标签依赖关系
To enable PTMGPT2 to identify critical sequence determinants essential for protein modifications, we designed a framework depicted in Fig. 2A that processes protein sequences to extract attention scores from the model’s last decoder layer. The attention mechanism is pivotal as it selectively weighs the importance of different segments of the input sequence during prediction. Particularly, the extracted attention scores from the final layer provided a granular view of the model’s focus across the input sequence. By aggregating the attention across 20 attention heads (AH) for each position in the sequence, PTMGPT2 revealed which amino acids or motifs the model deemed crucial in relation to the ‘POSITIVE’ token. The Position Specific Probability Matrix $(\mathsf{P S P M})^{19}$ , characterized by rows representing sequence positions and columns indicating amino acids, was a key output of this analysis. It sheds light on the proportional representation of each amino acid in the sequences, as weighted by the attention scores. PTMGPT2 thus offers a refined view of the probabilistic distribution of amino acid occurrences, revealing key patterns and preferences in amino acid positioning.
为使PTMGPT2能够识别蛋白质修饰所必需的关键序列决定因素,我们设计了图2A所示的框架,该框架处理蛋白质序列以从模型的最后一个解码器层提取注意力分数。注意力机制至关重要,因为它能在预测过程中选择性权衡输入序列不同片段的重要性。特别地,从最终层提取的注意力分数提供了模型在输入序列上关注焦度的细粒度视图。通过聚合序列中每个位置20个注意力头(AH)的注意力权重,PTMGPT2揭示了模型认为与"POSITIVE"标记相关的关键氨基酸或基序。位置特异性概率矩阵$(\mathsf{P S P M})^{19}$是该分析的关键输出,其行代表序列位置,列表示氨基酸。该矩阵通过注意力分数加权,阐明了各氨基酸在序列中的比例分布。PTMGPT2由此提供了氨基酸出现概率分布的精细化视图,揭示了氨基酸定位的关键模式与偏好。
Motifs $\mathbf{K}^{**}A^{*}A$ and $C^{**}K$ were identified in AH 10, while motifs $K^{\ast\ast\ast}\mathsf{K}^{\ast\ast\ast\ast}\mathsf{A}$ , $^{*}\mathrm{KH^{*}}$ , and ${\mathsf{K}}^{***}{\mathsf{K}}$ were detected in AH 7. In AH 19, motifs $\mathsf{K}^{*}\mathsf{C}$ and $\mathtt{C}^{*}\mathtt{K}$ motifs were observed, and the $\mathbf{*}\mathbf{G}\mathbf{K}^{*}$ motif was found in AH 15. Furthermore, motifs ${}^{*}\mathbf{E}\mathbf{K}^{*}$ and $\mathbf{K}\mathbf{L}^{**}\mathbf{E}\mathbf{R}$ were identified in AH 5, motifs $\mathsf{H}^{***}\mathsf{K}$ , $\mathbf{D}^{**}\mathbf{K},$ and ${}^{*}\mathsf{F K}^{*}$ were detected in $\mathsf{A H4}.$ The $\mathbf{A}^{**}\mathbf{K}$ motif was observed in AH13, $\mathbf{A}^{*}\mathbf{K}$ motif in AH2, and ${}^{*}\mathbf{KM}^{*}$ motif in AH 11. To validate the predictions made by PTMGPT2 for lysine (K) acetyl ation, as shown in Fig. 2B, we compared these with motifs identified in prior research that has undergone experimental validation. Expanding our analysis to protein kinase domains, we visualized motifs for the CMGC and AGC kinase families, as shown in Fig. 3A, B. Additionally, the motifs for the CAMK kinase family and general protein kinases are shown in Fig. 4A, B, respectively. The CMGC kinase family named after its main members, CDKs (cyclin-dependent kinases), MAPKs (mitogen-acti- vated protein kinases), GSKs (glycogen synthase kinases), and CDK-like kinases is involved in cell cycle regulation, signal transduction, and cellular differentiation 20. PTMGPT2 identified the common motif ${}^{*}{\mathsf{P}}^{*}{\mathsf{S P}}^{*}$ (Proline at positions $^{-2}$ and $^{+1}$ from the phosphor y late d serine residue) in this family. The AGC kinase family, comprising key serine/ threonine protein kinases such as PKA (protein kinase A), PKG (protein kinase G), and PKC (protein kinase C), plays a critical role in regulating metabolism, growth, proliferation, and survival21. The predicted common motif in this family was $\mathbf{R}^{**}\mathbf{S}\mathbf{L}$ (Arginine at position $^{-2}$ and leucine at position $^{+1}$ from either a phosphor y late d serine or threonine). The
在AH 10中识别到基序 $K^{**}\mathbf{A}^{*}\mathbf{A}$ 和 $\mathtt{C}^{**}\mathtt{K}$,而在AH 7中检测到基序 $\mathsf{K}^{\ast\ast\ast}\mathsf{K}^{\ast\ast\ast\ast}\mathsf{A}$、$^{*}\mathrm{KH^{*}}$ 和 ${\mathsf{K}}^{***}{\mathsf{K}}$。在AH 19中观察到基序 $\mathsf{K}^{*}\mathsf{C}$ 和 $\mathtt{C}^{*}\mathtt{K}$,在AH 15中发现基序 $\mathbf{*}\mathbf{G}\mathbf{K}^{*}$。此外,在AH 5中识别到基序 ${}^{*}\mathbf{E}\mathbf{K}^{*}$ 和 $\mathbf{K}\mathbf{L}^{**}\mathbf{E}\mathbf{R}$,在 $\mathsf{A H4}$ 中检测到基序 $\mathsf{H}^{***}\mathsf{K}$、$\mathbf{D}^{**}\mathbf{K},$ 和 ${}^{*}\mathsf{F K}^{*}$。在AH13中观察到基序 $\mathbf{A}^{**}\mathbf{K}$,在AH2中观察到基序 $\mathbf{A}^{*}\mathbf{K}$,在AH 11中观察到基序 ${}^{*}\mathbf{KM}^{*}$。为验证PTMGPT2对赖氨酸 (K) 乙酰化的预测(如图2B所示),我们将其与既往经过实验验证的研究中已识别的基序进行比较。将分析扩展至蛋白激酶结构域,我们可视化了CMGC和AGC激酶家族的基序(如图3A、B所示)。此外,CAMK激酶家族和通用蛋白激酶的基序分别显示在图4A、B中。CMGC激酶家族以其主要成员命名,包括CDKs(细胞周期蛋白依赖性激酶)、MAPKs(丝裂原活化蛋白激酶)、GSKs(糖原合成酶激酶)和CDK样激酶,参与细胞周期调控、信号转导和细胞分化[20]。PTMGPT2在该家族中识别出常见基序 ${}^{*}{\mathsf{P}}^{*}{\mathsf{S P}}^{*}$(在磷酸化丝氨酸残基的 $^{-2}$ 和 $^{+1}$ 位置为脯氨酸)。AGC激酶家族包含关键的丝氨酸/苏氨酸蛋白激酶,如PKA(蛋白激酶A)、PKG(蛋白激酶G)和PKC(蛋白激酶C),在调节代谢、生长、增殖和存活中起关键作用[21]。该家族预测的常见基序为 $\mathbf{R}^{**}\mathbf{S}\mathbf{L}$(在磷酸化丝氨酸或苏氨酸的 $^{-2}$ 位置为精氨酸,$^{+1}$ 位置为亮氨酸)。
Table 2 | Benchmark dataset results
| PTM | Model | MCC | F1Score | Precision | Recall |
| Succinylation (K) | LM-SuccSite34 | 43.94 | 62.26 | 53.05 | 45.34 |
| Psuc-EDBAM35 | 43.15 | 58.23 | 51.03 | 47.8 | |
| SuccinSite36 | 15.87 | 34.47 | 64.95 | 23.46 | |
| Deep SuccinylSite37 | 39.77 | 59.4 | 70.51 | 48.32 | |
| Deep-KSuccSite38 | 36.88 | 55.89 | 47.41 | 48.06 | |
| Succ-PTMGPT2 | 51.88 | 60.74 | 79.15 | 49.27 | |
| Sumoylation (K) | Musite-Web39 | 42.84 | 41.73 | 63.96 | 27.21 |
| CapsNet40 | 37.05 | 33.35 | 68.75 | 20.45 | |
| ResSum041 | 21.07 | 25.59 | 15.56 | 61.97 | |
| GPS Sum042 | 61.02 | 65.76 | 70.38 | 25.67 | |
| Sumoy-PTMGPT2 | 66.93 | 69.21 | 75.87 | 63.63 | |
| N-linked Glycosylation (N) | Musite-Web39 | 65.30 | 68.56 | 61.42 | 87.08 |
| CapsNet40 | 50.38 | 56.18 | 66.04 | 45.65 | |
| LMNglyPred43 | 39.34 | 35.17 | |||
| N-linked-PTMGPT2 | 70.92 | 74.21 | 57.49 64.35 | 14.58 87.65 | |
| Methyl Arginine (R) | Musite-Web39 | 67.77 | 68.99 | 85.41 | 57.86 |
| CapsNet40 | 65.39 | 65.12 | 90.03 | 51.08 | |
| DeepRMethylSite44 | 34.02 | 52.49 | 79.89 | 39.08 | |
| PRmePred45 | 53.76 | 54.33 | 76.45 | 36.22 | |
| CNNArginineMe46 | 34.55 | 36.98 | 97.42 | 22.82 | |
| Lysine Acetylation (K) | RMethyl-PTMGPT2 | 80.51 | 81.32 | 95.14 | 71.01 |
| Musite-Web39 | 21.63 | 36.14 | 94.86 | 22.32 | |
| CapsNet40 | 21.62 | 36.54 | 93.46 | ||
| GPS-PAIL47 | 12.35 | 16.14 | 72.63 | 22.71 9.07 | |
| Ubiquitination (K) | KAcetyl-PTMGPT2 | 22.09 | 40.51 | 96.06 | 25.67 |
| Musite-Web39 | 26.24 | 27.67 | 75.67 | 16.93 | |
| DL-Ubiq48 | 10.91 | 33.37 | |||
| Ubiq-PTMGPT2 | 31.25 | 35.74 | 36.67 80.46 | 23.76 22.97 | |
| O-linked-Glycosylation (S,T) | Musite-Web39 | 49.89 | 50.29 | 61.64 | 40.38 |
| OGlyThr49 | 41.6 | 50.71 | 52.82 | 60.78 | |
| GlyCopp50 | 41.05 | 14.38 | |||
| O-linked-PTMGPT2 | 58.97 | 61.80 | 50.85 | 46.67 65.14 | |
| S-Nitrosylation (C) | PCysMod51 | 48.53 | 66.67 | 58.79 62.50 | 71.42 |
| DeepNitro52 | 79.55 | 45.45 | |||
| PresSNO53 | 59.45 | 62.50 74.21 | 72.72 | ||
| pIMSNOSite54 | 69.52 | 84.19 | |||
| SNitro-PTMGPT2 | 55.91 | 17.60 | 14.74 | 21.82 | |
| Malonylation (K) | DL-Malosite55 | 73.61 69.78 | 80.49 77.12 | 84.30 | 77.01 81.94 |
| Methyl Lysine (K) | Maloy-PTMGPT2 | 73.03 | 78.14 | 72.83 82.95 | 73.85 |
| Musite-Web39 | 14.97 | 10.13 | 72.44 | 5.44 | |
| MethylSite56 | 38.60 | 35.54 | 39.41 | 66.66 | |
| KMethyl-PTMGPT2 | 41.07 | 36.35 | 88.68 | 22.86 | |
| Phosphorylation (S, T) | Musite-Web39 | 17.73 | 12.22 | 6.67 | 72.74 |
| CapsNet40 | 6.71 | 71.94 | |||
| PhosphoST-PTMGPT2 | 16.23 16.37 | 12.28 15.35 | 9.14 | 46.97 | |
| Musite-Web39 | 40.83 | 43.03 | 29.82 | 77.27 | |
| Phosphorylation (Y) | CapsNet40 | 43.85 | 46.61 | 36.58 | 68.18 |
| PhosphoY-PTMGPT2 | 48.83 | 46.98 | 30.95 | 97.51 | |
| DeepGSH57 | 70.77 | 75.85 | |||
| Glutathio-PTMGPT2 | 74.23 | 75.28 | |||
| ProtTrans-Glutar58 | 78.28 | 82.37 | 90.07 | 75.89 | |
| Glutarylation (K) | 62.99 | 59.45 | 68.09 | 79.16 | |
| Glutary-PTMGPT2 PrAS59 | 69.47 76.00 | 73.78 NR | 71.02 NR | 76.76 81.2 | |
| Ami-PTMGPT2 | 80.78 | 76.59 | 66.67 | 86.76 | |
| Musite-Web39 | 35.69 | 47.82 | 79.78 | 49.45 |
表 2 | 基准数据集结果
| PTM | 模型 | MCC | F1分数 | 精确率 | 召回率 |
|---|---|---|---|---|---|
| 琥珀酰化 (K) | |||||
| LM-SuccSite[34] | 43.94 | 62.26 | 53.05 | 45.34 | |
| Psuc-EDBAM[35] | 43.15 | 58.23 | 51.03 | 47.8 | |
| SuccinSite[36] | 15.87 | 34.47 | 64.95 | 23.46 | |
| Deep SuccinylSite[37] | 39.77 | 59.4 | 70.51 | 48.32 | |
| Deep-KSuccSite[38] | 36.88 | 55.89 | 47.41 | 48.06 | |
| Succ-PTMGPT2 | 51.88 | 60.74 | 79.15 | 49.27 | |
| 苏素化 (K) | |||||
| Musite-Web[39] | 42.84 | 41.73 | 63.96 | 27.21 | |
| CapsNet[40] | 37.05 | 33.35 | 68.75 | 20.45 | |
| ResSumo[41] | 21.07 | 25.59 | 15.56 | 61.97 | |
| GPS Sumo[42] | 61.02 | 65.76 | 70.38 | 25.67 | |
| Sumoy-PTMGPT2 | 66.93 | 69.21 | 75.87 | 63.63 | |
| N-连接糖基化 (N) | |||||
| Musite-Web[39] | 65.30 | 68.56 | 61.42 | 87.08 | |
| CapsNet[40] | 50.38 | 56.18 | 66.04 | 45.65 | |
| LMNglyPred[43] | 39.34 | 35.17 | |||
| N-linked-PTMGPT2 | 70.92 | 74.21 | 57.49 64.35 | 14.58 87.65 | |
| 精氨酸甲基化 (R) | |||||
| Musite-Web[39] | 67.77 | 68.99 | 85.41 | 57.86 | |
| CapsNet[40] | 65.39 | 65.12 | 90.03 | 51.08 | |
| DeepRMethylSite[44] | 34.02 | 52.49 | 79.89 | 39.08 | |
| PRmePred[45] | 53.76 | 54.33 | 76.45 | 36.22 | |
| CNNArginineMe[46] | 34.55 | 36.98 | 97.42 | 22.82 | |
| RMethyl-PTMGPT2 | 80.51 | 81.32 | 95.14 | 71.01 | |
| 赖氨酸乙酰化 (K) | |||||
| Musite-Web[39] | 21.63 | 36.14 | 94.86 | 22.32 | |
| CapsNet[40] | 21.62 | 36.54 | 93.46 | ||
| GPS-PAIL[47] | 12.35 | 16.14 | 72.63 | 22.71 9.07 | |
| KAcetyl-PTMGPT2 | 22.09 | 40.51 | 96.06 | 25.67 | |
| 泛素化 (K) | |||||
| Musite-Web[39] | 26.24 | 27.67 | 75.67 | 16.93 | |
| DL-Ubiq[48] | 10.91 | 33.37 | |||
| Ubiq-PTMGPT2 | 31.25 | 35.74 | 36.67 80.46 | 23.76 22.97 | |
| O-连接糖基化 (S,T) | |||||
| Musite-Web[39] | 49.89 | 50.29 | 61.64 | 40.38 | |
| OGlyThr[49] | 41.6 | 50.71 | 52.82 | 60.78 | |
| GlyCopp[50] | 41.05 | 14.38 | |||
| O-linked-PTMGPT2 | 58.97 | 61.80 | 50.85 | 46.67 65.14 | |
| S-亚硝基化 (C) | |||||
| PCysMod[51] | 48.53 | 66.67 | 58.79 62.50 | 71.42 | |
| DeepNitro[52] | 79.55 | 45.45 | |||
| PresSNO[53] | 59.45 | 62.50 74.21 | 72.72 | ||
| pIMSNOSite[54] | 69.52 | 84.19 | |||
| SNitro-PTMGPT2 | 55.91 | 17.60 | 14.74 | 21.82 | |
| 丙二酰化 (K) | |||||
| DL-Malosite[55] | 73.61 69.78 | 80.49 77.12 | 84.30 | 77.01 81.94 | |
| Maloy-PTMGPT2 | 73.03 | 78.14 | 72.83 82.95 | 73.85 | |
| 赖氨酸甲基化 (K) | |||||
| Musite-Web[39] | 14.97 | 10.13 | 72.44 | 5.44 | |
| MethylSite[56] | 38.60 | 35.54 | 39.41 | 66.66 | |
| KMethyl-PTMGPT2 | 41.07 | 36.35 | 88.68 | 22.86 | |
| 磷酸化 (S, T) | |||||
| Musite-Web[39] | 17.73 | 12.22 | 6.67 | 72.74 | |
| CapsNet[40] | 6.71 | 71.94 | |||
| PhosphoST-PTMGPT2 | 16.23 16.37 | 12.28 15.35 | 9.14 | 46.97 | |
| Musite-Web[39] | 40.83 | 43.03 | 29.82 | 77.27 | |
| 磷酸化 (Y) | |||||
| CapsNet[40] | 43.85 | 46.61 | 36.58 | 68.18 | |
| PhosphoY-PTMGPT2 | 48.83 | 46.98 | 30.95 | 97.51 | |
| 谷胱甘肽化 | |||||
| DeepGSH[57] | 70.77 | 75.85 | |||
| Glutathio-PTMGPT2 | 74.23 | 75.28 | |||
| ProtTrans-Glutar[58] | 78.28 | 82.37 | 90.07 | 75.89 | |
| 戊二酰化 (K) | |||||
| 62.99 | 59.45 | 68.09 | 79.16 | ||
| Glutary-PTMGPT2 | 69.47 76.00 | 73.78 NR | 71.02 NR | 76.76 81.2 | |
| 酰胺化 | |||||
| Ami-PTMGPT2 | 80.78 | 76.59 | 66.67 | 86.76 | |
| Musite-Web[39] | 35.69 | 47.82 | 79.78 | 49.45 |
Table 2 (continued) | Benchmark dataset results
| PTM | Model MCC | F1Score | Precision | Recall |
| CapsNet40 | 39.81 47.67 | 73.89 | 43.94 | |
| GPS-Palm60 | 24.05 28.49 | 16.69 | 97.19 | |
| Palm-PTMGPT2 | 41.37 | 48.34 | 42.73 55.64 | |
| Proline Hydroxylation (P) | Musite-Web39 | 78.08 | 80.58 98.32 | 67.47 |
| CapsNet40 | 78.87 | 85.92 | 94.66 74.79 | |
| ProHydroxy-PTMGPT2 | 89.89 | 92.30 96.18 | 88.73 | |
| Lysine Hydroxylation (K) | Musite-Web39 | 57.67 63.76 | 76.33 | 72.12 |
| CapsNet40 | 58.87 | 65.92 | 74.66 | |
| LysHydroxy-PTMGPT2 | 66.45 | 68.18 88.23 | 74.79 55.56 | |
| Formylation (K) | CapsNet40 | 40.19 33.33 | 24.09 | 68.34 |
| Musite-Web39 | 39.55 | 28.47 | 23.24 | |
| Formy-PTMGPT2 | 44.58 39.97 | 26.92 | 88.88 77.78 |
Top values for each PTM are represented in bold
表 2 (续表) | 基准数据集结果
| PTM | 模型 MCC | F1分数 | 精确率 | 召回率 |
|---|---|---|---|---|
| CapsNet40 | 39.81 47.67 | 73.89 | 43.94 | |
| GPS-Palm60 | 24.05 28.49 | 16.69 | 97.19 | |
| Palm-PTMGPT2 | 41.37 | 48.34 | 42.73 55.64 | |
| 脯氨酸羟基化 (P) | Musite-Web39 | 78.08 | 80.58 98.32 | 67.47 |
| CapsNet40 | 78.87 | 85.92 | 94.66 74.79 | |
| ProHydroxy-PTMGPT2 | 89.89 | 92.30 96.18 | 88.73 | |
| 赖氨酸羟基化 (K) | Musite-Web39 | 57.67 63.76 | 76.33 | 72.12 |
| CapsNet40 | 58.87 | 65.92 | 74.66 | |
| LysHydroxy-PTMGPT2 | 66.45 | 68.18 88.23 | 74.79 55.56 | |
| 甲酰化 (K) | CapsNet40 | 40.19 33.33 | 24.09 | 68.34 |
| Musite-Web39 | 39.55 | 28.47 | 23.24 | |
| Formy-PTMGPT2 | 44.58 39.97 | 26.92 | 88.88 77.78 |
各PTM类型的最高值以粗体标出
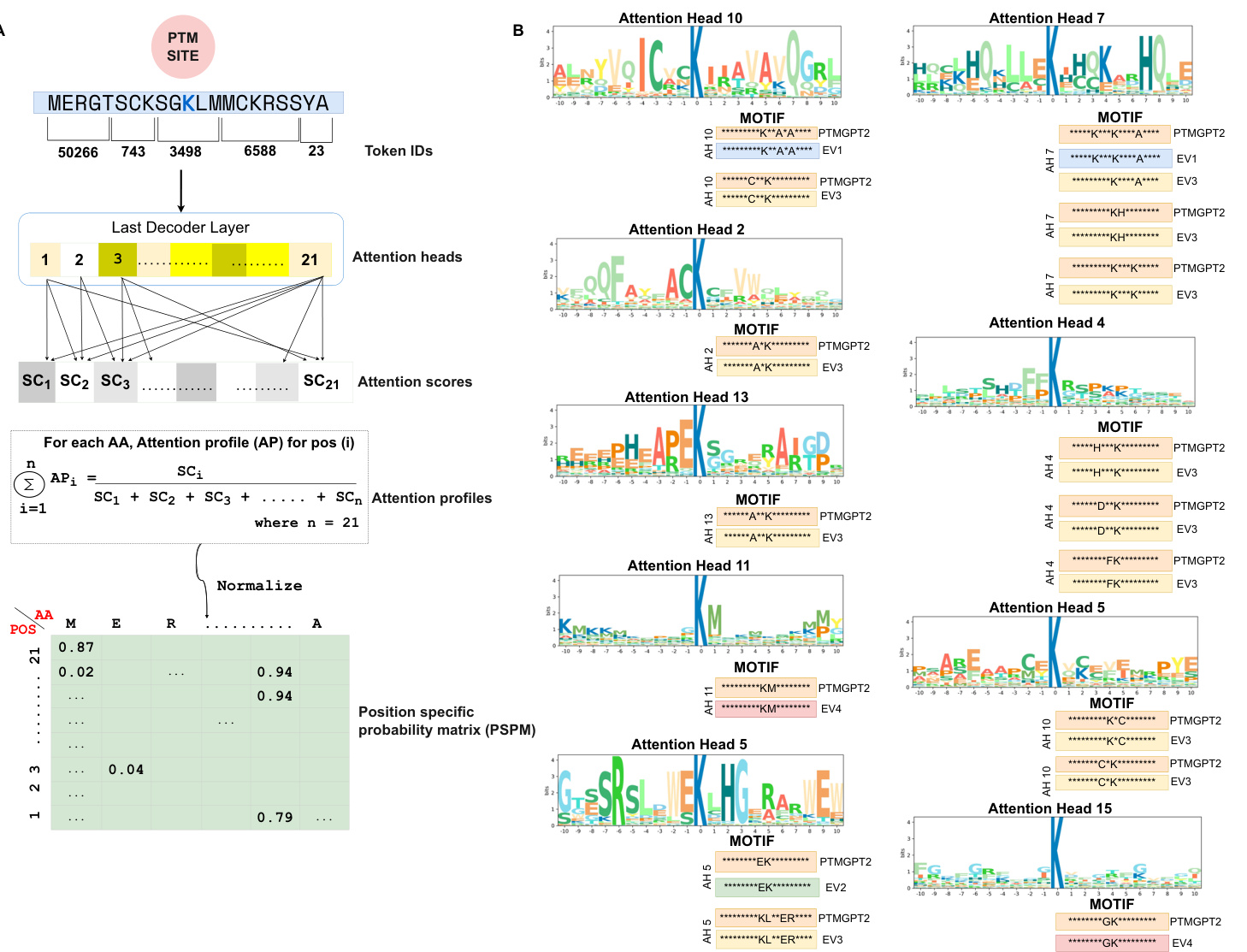
Fig. 2 | Attention head analysis of lysine (K) acetyl ation by PTMGPT2. A Computation of attention scores from the model’s last decoder layer, detailing the process of generating a Position-Specific Probability Matrix (PSPM) for a targeted protein sequence.’SC’ denotes attention scores,’AP’ denotes attention

图 2 | PTMGPT2 对赖氨酸 (K) 乙酰化的注意力头分析。A 从模型最后一个解码器层计算注意力分数,详细说明为目标蛋白质序列生成位置特异性概率矩阵 (PSPM) 的过程。'SC' 表示注意力分数,'AP' 表示注意力概率。
CAMK kinase family, which includes key members like CaMK2 and CAMKL, is crucial in signaling pathways related to neurological disorders, cardiac diseases, and other conditions associated with calcium signaling dys regulation 22. The common motif identified by PTMGPT2 in CAMK was ${\bf R}^{\ast\ast}{\sf S}$ (Arginine at position $^{-2}$ from either a profiles,’AA’ represents an amino acid, and’n’ is the number of amino acids in a sub sequence. B Sequence motifs validated by experimentally verified studies— $\mathrm{EVI}^{61}$ , $\mathrm{EV}2^{62}$ , $\mathrm{EV}3^{63}$ , $\mathrm{EV}4^{47}$ .’AH’ denotes attention head.
CAMK激酶家族, 包括CaMK2和CAMKL等关键成员, 在与神经系统疾病、心脏病及其他钙信号传导失调相关的信号通路中至关重要 22。PTMGPT2在CAMK中识别出的共同基序是 ${\bf R}^{\ast\ast}{\sf S}$ (精氨酸位于 $^{-2}$ 位点)。"AA"代表氨基酸,"n"表示子序列中的氨基酸数量。B 经实验验证研究确认的序列基序—— $\mathrm{EVI}^{61}$ , $\mathrm{EV}2^{62}$ , $\mathrm{EV}3^{63}$ , $\mathrm{EV}4^{47}$ 。"AH"表示注意力头。
phosphor y late d serine or threonine). Further analysis of general protein kinases revealed distinct patterns: DMPK kinase exhibited the motif $\mathbf{RR}^{*}\mathbf{T}$ (Arginine at positions $^{-2}$ and $-3,$ ), MAPKAPK kinase followed the $\mathtt{R}^{*}\mathtt{L S}$ motif (Arginine at position $^{-3}$ and leucine at position −1), AKT kinase was characterized by the ${\tt R}^{*}{\tt R S}$ motif (Arginine at positions $^{-1}$ and $^{-3}$ ), CK1 kinase showed $\mathsf{K}^{*}\mathsf{K}^{**}\mathsf{S}/\mathsf{T}$ (Lysine at positions $^{-3}$ and $-5.$ ), and CK2 kinase was defined by the $\mathsf{S D}^{*}\mathbf{E}$ motif (Aspartate at position $^{+1}$ and glutamate at position $^{+3}$ ). These comparisons underscored PTMGPT2’s ability to accurately identify motifs associated with diverse kinase groups and PTM types. PSPM matrices, corresponding to 20 attention heads across all 19 PTM types, are detailed in Supplementary Data 1. These insights are crucial for deciphering the intricate mechanisms underlying protein modifications. Consequently, this analysis, driven by the PTMGPT2 model, forms a core component of our exploration into the contextual relationships between protein sequences and their predictive labels.
磷酸化的丝氨酸或苏氨酸)。对通用蛋白激酶的进一步分析揭示了不同的模式:DMPK激酶呈现基序 $\mathbf{RR}^{*}\mathbf{T}$(精氨酸位于 $^{-2}$ 和 $-3$ 位),MAPKAPK激酶遵循 $\mathtt{R}^{*}\mathtt{L S}$ 基序(精氨酸位于 $^{-3}$ 位,亮氨酸位于-1位),AKT激酶以 ${\tt R}^{*}{\tt R S}$ 基序为特征(精氨酸位于 $^{-1}$ 和 $^{-3}$ 位),CK1激酶显示 $\mathsf{K}^{*}\mathsf{K}^{**}\mathsf{S}/\mathsf{T}$(赖氨酸位于 $^{-3}$ 和 $-5$ 位),CK2激酶则由 $\mathsf{S D}^{*}\mathbf{E}$ 基序定义(天冬氨酸位于 $^{+1}$ 位,谷氨酸位于 $^{+3}$ 位)。这些比较结果突显了PTMGPT2准确识别不同激酶组和PTM类型相关基序的能力。对应所有19种PTM类型中20个注意力头的PSPM矩阵详见补充数据1。这些发现对解析蛋白质修饰的复杂机制至关重要。因此,这项由PTMGPT2模型驱动的分析构成了我们探索蛋白质序列与其预测标签间上下文关系的核心组成部分。
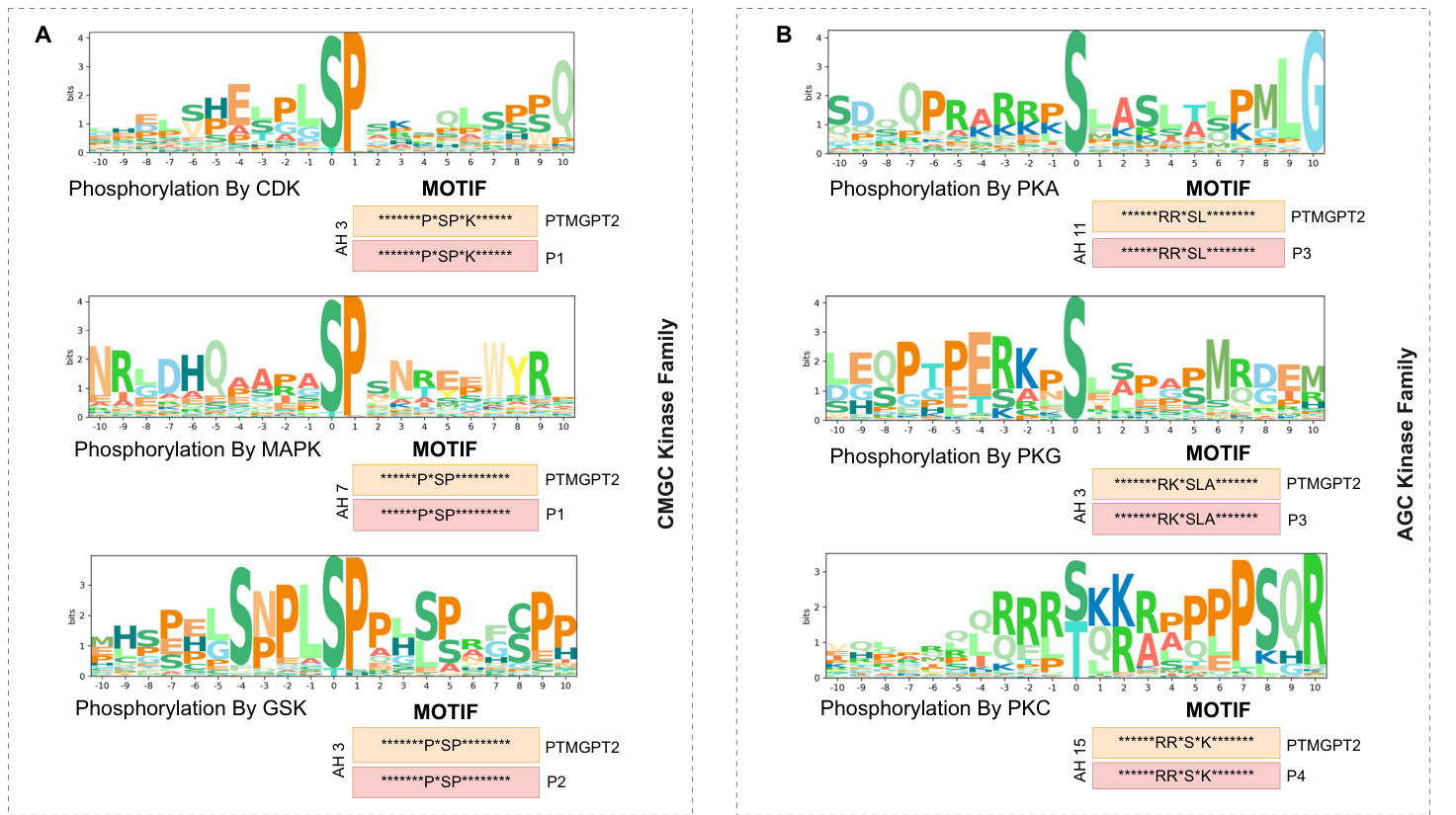
Fig. 3 | Attention head analysis of the CMGC kinase family and the AGC kinase family by PTMGPT2. A Motifs from CMGC kinase family validated against ${\mathsf{P}}1^{64}$ and ${\mathsf{P}}2^{65}$ . B AGC kinase family motifs validated against $\mathsf{P}3^{66}$ and $\mathbf{P}4^{67}$ . The’P*SP’ motif is
图 3 | PTMGPT2对CMGC激酶家族和AGC激酶家族的注意力头分析。A 基于 ${\mathsf{P}}1^{64}$ 和 ${\mathsf{P}}2^{65}$ 验证的CMGC激酶家族基序。B 基于 $\mathsf{P}3^{66}$ 和 $\mathbf{P}4^{67}$ 验证的AGC激酶家族基序。"P*SP"基序是
Recent uniprot entries validate PTMGPT2’s robust generalization abilities
最新UniProt条目验证PTMGPT2的稳健泛化能力
To demonstrate PTMGPT2’s robust predictive capabilities on unseen datasets, we extracted proteins recently released on UniProt, strictly selecting those added after June 1, 2023, to validate the model’s performance. We ensured these proteins were not present in the training or benchmark datasets from DBPTM (version May 2023), which was a crucial step in the validation process. A total of 31 proteins that met our criteria were identified, associated with PTMs such as phosphor yl ation (S, T, Y), methyl ation (K), and acetyl ation (K). The accurate prediction of PTMs in recently identified proteins not only validates the effectiveness of our model but also underscores its potential to advance research in protein biology and PTM site identification. These predictions are pivotal for pinpointing the precise locations and characteristics of modifications within the protein sequences, which are crucial for verifying PTMGPT2’s performance. The predictions for all 31 proteins, along with the ground truth, are detailed in Supplementary Table S1−S5.
为验证PTMGPT2在未知数据集上的强大预测能力,我们从UniProt中提取了近期发布的蛋白质,严格筛选2023年6月1日后新增的蛋白质以评估模型性能。我们确保这些蛋白质未出现在DBPTM(2023年5月版)的训练集或基准数据集中,这是验证过程中的关键步骤。最终确定了31个符合标准的蛋白质,涉及磷酸化(S, T, Y)、甲基化(K)和乙酰化(K)等翻译后修饰。对新发现蛋白质中PTM的准确预测不仅验证了我们模型的有效性,更凸显了其在推动蛋白质生物学研究和PTM位点识别方面的潜力。这些预测对于精确定位蛋白质序列中修饰的具体位置和特征至关重要,为验证PTMGPT2的性能提供了关键依据。所有31个蛋白质的预测结果及真实数据详见补充表S1-S5。
PTMGPT2 identifies mutation hotspots in ph os ph o sites of TP53, BRAF, and RAF1 genes
PTMGPT2 识别 TP53、BRAF 和 RAF1 基因磷酸化位点的突变热点
Protein PTMs play a vital role in regulating protein function. A key aspect of PTMs is their interplay with mutations, particularly near common in the CMGC kinase family, whereas the’R**S’ motif is common in the AGC kinase family.’AH’ denotes attention head.
蛋白质翻译后修饰 (PTMs) 在调控蛋白质功能中起着至关重要的作用。PTMs 的一个关键方面是其与突变的相互作用,特别是在 CMGC 激酶家族中常见的 (R**S) 基序,而 (R**S) 基序在 AGC 激酶家族中常见。(AH) 表示注意力头。
modification sites, where mutations can significantly impact protein function and potentially lead to disease. Previous studies23–25 indicate a strong correlation between pathogenic mutations and proximity to ph os ph os erin e sites, with over $70%$ of PTM-related mutations occurring in phosphor yl ation regions. Therefore, our study primarily targets ph os ph os erin e sites to provide a more in-depth understanding of PTM-related mutations. This study aims to evaluate PTMGPT2’s ability to identify mutations within 1−8 residues flanking a ph os ph os erin e site, without explicit mutation site annotations during training. For this, we utilized the dbSNP database26, which includes information on human single nucleotide variations linked to both common and clinical mutations. $T P53^{27}$ is a critical tumor suppressor gene, with mutations in TP53 being among the most prevalent in human cancers. When mutated, TP53 may lose its tumor-suppressing function, leading to unc
