GeneGPT: Augmenting Large Language Models with Domain Tools for Improved Access to Biomedical Information
GeneGPT:通过领域工具增强大语言模型以提升生物医学信息获取能力
Qiao $\mathbf{Jin^{ * }}$ , Yifan $\mathbf{Yang^{\pm0}}$ , Qingyu Chen♣, Zhiyong Lu♣
Qiao $\mathbf{Jin^{ * }}$,Yifan $\mathbf{Yang^{\pm0}}$,Qingyu Chen♣,Zhiyong Lu♣
♣ National Library of Medicine, National Institutes of Health ♥ University of Maryland, College Park {qiao.jin, yifan.yang3, qingyu.chen, zhiyong.lu}@nih.gov
♣ 美国国立卫生研究院国家医学图书馆 ♥ 马里兰大学帕克分校 {qiao.jin, yifan.yang3, qingyu.chen, zhiyong.lu}@nih.gov
Abstract
摘要
While large language models (LLMs) have been successfully applied to various tasks, they still face challenges with hallucinations. Augmenting LLMs with domain-specific tools such as database utilities can facilitate easier and more precise access to specialized knowledge. In this paper, we present GeneGPT, a novel method for teaching LLMs to use the Web APIs of the National Center for Biotechnology Information (NCBI) for answering genomics questions. Specifically, we prompt Codex to solve the GeneTuring tests with NCBI Web APIs by in-context learning and an augmented decoding algorithm that can detect and execute API calls. Experimental results show that GeneGPT achieves state-of-theart performance on eight tasks in the GeneTuring benchmark with an average score of 0.83, largely surpassing retrieval-augmented LLMs such as the new Bing (0.44), biomedical LLMs such as BioMedLM (0.08) and BioGPT (0.04), as well as GPT-3 (0.16) and ChatGPT (0.12). Our further analyses suggest that: (1) API demonstrations have good cross-task generalizability and are more useful than documentations for in-context learning; (2) GeneGPT can generalize to longer chains of API calls and answer multi-hop questions in GeneHop, a novel dataset introduced in this work; (3) Different types of errors are enriched in different tasks, providing valuable insights for future improvements.
虽然大语言模型(LLM)已成功应用于各种任务,但仍面临幻觉问题。通过整合领域专用工具(如数据库实用程序)可以增强大语言模型,使其更便捷精确地获取专业知识。本文提出GeneGPT——一种教导大语言模型使用美国国家生物技术信息中心(NCBI)网络API回答基因组学问题的新方法。具体而言,我们通过上下文学习提示Codex解决GeneTuring测试,并采用能检测和执行API调用的增强解码算法。实验结果表明,GeneGPT在GeneTuring基准测试的八个任务中平均得分0.83,显著超越检索增强型大语言模型如新版必应(0.44)、生物医学大语言模型如BioMedLM(0.08)和BioGPT(0.04),以及GPT-3(0.16)和ChatGPT(0.12)。进一步分析表明:(1) API演示具有跨任务泛化能力,其上下文学习效果优于文档说明;(2) GeneGPT能泛化至更长的API调用链,并回答本工作新数据集GeneHop中的多跳问题;(3) 不同任务中富集的错误类型各异,为未来改进提供了宝贵洞见。
1 Introduction
1 引言
Large language models (LLMs) such as PaLM (Chowdhery et al., 2022) and GPT-4 (OpenAI, 2023) have shown great success on a wide range of general-domain Natural Language Processing (NLP) tasks. They also achieve state-of-the-art (SOTA) performance on domain-specific tasks like biomedical question answering (Singhal et al., 2022; Liévin et al., 2022; Nori et al., 2023). How- ever, since there is no intrinsic mechanism for autoregressive LLMs to “consult” with any source of truth, they can generate plausible-sounding but incorrect content (Ji et al., 2023). To tackle the hallucination issue, various studies have been proposed to augment LLMs (Mialon et al., 2023) by either conditioning them on retrieved relevant content (Guu et al., 2020; Lewis et al., 2020; Borgeaud et al., 2022) or allowing them to use other external tools such as program APIs (Gao et al., 2022; Parisi et al., 2022; Schick et al., 2023; Qin et al., 2023).
大语言模型 (LLM) 如 PaLM (Chowdhery 等人, 2022) 和 GPT-4 (OpenAI, 2023) 在广泛通用领域的自然语言处理 (NLP) 任务中取得了巨大成功。它们也在生物医学问答等特定领域任务中实现了最先进 (SOTA) 性能 (Singhal 等人, 2022; Liévin 等人, 2022; Nori 等人, 2023)。然而,由于自回归大语言模型缺乏内在机制来"查阅"真实信息来源,它们可能生成听起来合理但实际错误的内容 (Ji 等人, 2023)。为解决幻觉问题,各种研究提出通过两种方式增强大语言模型 (Mialon 等人, 2023):基于检索相关内容进行条件生成 (Guu 等人, 2020; Lewis 等人, 2020; Borgeaud 等人, 2022),或允许其使用程序 API 等其他外部工具 (Gao 等人, 2022; Parisi 等人, 2022; Schick 等人, 2023; Qin 等人, 2023)。
In this work, we propose to teach LLMs to use the Web APIs of the National Center for Biotechnology Information (NCBI). NCBI provides API access to its entire biomedical databases and tools, including Entrez Programming Utilities (E-utils) and Basic Local Alignment Search Tool (BLAST) URL API (Altschul et al., 1990; Schuler et al., 1996; Sayers et al., 2019). Enabling LLMs to use NCBI Web APIs can provide easier and more pre- cise access to biomedical information, especially for users who are inexperienced with the database systems. More importantly, Web APIs can relieve users from locally implementing functionalities, maintaining large databases, and heavy computation burdens because the only requirement for using Web APIs is an internet connection.
在本研究中,我们提出教授大语言模型使用美国国家生物技术信息中心(NCBI)的Web API。NCBI通过API开放其全部生物医学数据库和工具,包括Entrez编程实用程序(E-utils)和基本局部比对搜索工具(BLAST) URL API (Altschul et al., 1990; Schuler et al., 1996; Sayers et al., 2019)。使大语言模型能够调用NCBI Web API可为用户提供更便捷、精准的生物医学信息访问途径,尤其有利于不熟悉该数据库系统的用户。更重要的是,Web API能免除用户本地实现功能、维护大型数据库和承担繁重计算任务的负担,因为使用Web API的唯一要求就是互联网连接。
We introduce GeneGPT, a novel method that prompts Codex (Chen et al., 2021) to use NCBI Web APIs by in-context learning (Brown et al., 2020). GeneGPT consists of two main modules: (a) a specifically designed prompt that consists of documentation s and demonstrations of API usage, and (b) an inference algorithm that integrates API calls in the Codex decoding process. We evaluate GeneGPT on GeneTuring (Hou and Ji, 2023), a question answering (QA) benchmark for genomics, and compare GeneGPT to a variety of other LLMs such as the new ${\mathrm{Bing}}^{1}$ , ChatGPT2, and BioGPT (Luo et al., 2022). GeneGPT achieves the best performance on eight GeneTuring tasks with an average score of 0.83, which is remarkably higher than the previous SOTA (0.44 by New Bing). In addition, we systematically characterize GeneGPT and find that: (1) API demonstrations are more useful than documentation s for in-context learning; (2) GeneGPT generalizes to longer chains of sub question decomposition and API calls with simple demonstrations; (3) GeneGPT makes specific errors that are enriched for each task.
我们介绍GeneGPT,这是一种通过上下文学习(Brown等人,2020)引导Codex(Chen等人,2021)使用NCBI Web API的新方法。GeneGPT包含两个主要模块:(a) 由API使用文档和演示组成的特制提示词;(b) 在Codex解码过程中整合API调用的推理算法。我们在基因组学问答基准GeneTuring(Hou和Ji,2023)上评估GeneGPT,并将其与多种大语言模型进行比较,如新版${\mathrm{Bing}}^{1}$、ChatGPT2和BioGPT(Luo等人,2022)。GeneGPT在八项GeneTuring任务中以0.83的平均分取得最佳性能,显著优于之前的SOTA(New Bing的0.44分)。此外,我们系统分析了GeneGPT的特性发现:(1) 对于上下文学习,API演示比文档更有用;(2) 通过简单演示,GeneGPT能泛化至更长的子问题分解和API调用链;(3) GeneGPT会产生特定于每项任务的错误类型。

Figure 1: Left: GeneGPT uses NCBI Web API documentation s and demonstrations in the prompt for in-context learning. Right: Examples of GeneGPT answering GeneTuring and GeneHop questions with NCBI Web APIs.
图 1: 左图: GeneGPT 在提示中使用 NCBI Web API 文档和演示进行上下文学习。右图: GeneGPT 使用 NCBI Web API 回答 GeneTuring 和 GeneHop 问题的示例。
In summary, our contributions are three-fold:
总之,我们的贡献体现在三个方面:
- We introduce GeneGPT, a novel method that uses NCBI Web APIs to answer biomedical questions. To the best of our knowledge, this is the first study on augmenting LLMs with domain-specific Web API tools.
- 我们提出GeneGPT,这是一种利用NCBI Web API解答生物医学问题的新方法。据我们所知,这是首个通过领域专用Web API工具增强大语言模型的研究。
- GeneGPT achieves SOTA performance on 8 tasks in the GeneTuring benchmark, largely outperforming previous best results by $88%$ (0.83 v.s. 0.44 set by the new Bing).
- GeneGPT在GeneTuring基准测试的8项任务中实现了SOTA(当前最优)性能,以88%的显著优势超越此前由新版Bing保持的最佳结果(0.83 vs 0.44)。
- We conduct experiments to further characterize GeneGPT, including ablation, probing, and error analyses. We also contribute a novel GeneHop dataset, and use it to show that GeneGPT can perform chain-of-thought API calls to answer multi-hop genomics questions.
- 我们通过消融实验、探测分析和错误分析进一步验证GeneGPT的特性,并贡献了新型GeneHop数据集,证明GeneGPT能通过思维链式API调用回答多跳基因组学问题。
2 GeneGPT
2 GeneGPT
In this section, we first introduce the general functions and syntax of NCBI Web APIs (§2.1). We then describe two key components of GeneGPT: its prompt design for in-context learning (§2.2) and the inference algorithm (§2.3).
在本节中,我们首先介绍NCBI Web API的基本功能和语法(§2.1),随后阐述GeneGPT的两个核心组件:面向上下文学习的提示设计(§2.2)以及推理算法(§2.3)。
2.1 NCBI Web APIs
2.1 NCBI Web APIs
We utilize NCBI Web APIs of E-utils3 that provide access to biomedical databases and the BLAST tool4 for DNA sequence alignment. Web API calls are implemented by the urllib library in Python.
我们利用NCBI的E-utils3网络API访问生物医学数据库,并使用BLAST工具4进行DNA序列比对。网络API调用通过Python语言的urllib库实现。
E-utils. It is the API for accessing the Entrez portal (Schuler et al., 1996), which is a system that covers 38 NCBI databases of biomedical data such as genes and proteins (Sayers et al., 2019). The E-utils API provides a fixed URL syntax for rapidly retrieving such biomedical information. Specifically, the base URL for an E-utils request is “https://eutils.ncbi.nlm.nih.gov/ entrez/eutils/{function}.fcgi”, where function can be esearch, efetch, or esummary. Typically, the user first calls esearch to get the unique database identifiers of a given query term. Then, efetch or esummary can be called to get the full records or text summaries of a given list of identifiers returned by esearch. Important arguments in the URL request include the search term or ids (term or id), the database to use (db), the maximum number of returned items (retmax), and the return format (retmode).
E-utils。这是用于访问Entrez门户的API (Schuler et al., 1996),该系统覆盖了38个NCBI生物医学数据库,如基因和蛋白质数据 (Sayers et al., 2019)。E-utils API提供了固定的URL语法用于快速检索此类生物医学信息。具体而言,E-utils请求的基础URL为"https://eutils.ncbi.nlm.nih.gov/entrez/eutils/{function}.fcgi",其中function可以是esearch、efetch或esummary。通常,用户首先调用esearch获取给定查询词的唯一数据库标识符,随后通过efetch或esummary获取由esearch返回的标识符列表对应的完整记录或文本摘要。URL请求中的重要参数包括搜索词或标识符(term或id)、使用的数据库(db)、返回条目最大数量(retmax)以及返回格式(retmode)。
BLAST URL API. BLAST takes as input a sequence of nucleotides or amino acids and finds the most similar sequences in the database (Altschul et al., 1990; Boratyn et al., 2013). The re- sults can be used to infer relationships between sequences or identify members of gene families. The BLAST API allows users to submit queries to find regions of similarities be- tween nucleotide or protein sequences to existing databases using the BLAST algorithm on NCBI servers. The base URL for the BLAST URL API is “https://blast.ncbi.nlm.nih.gov/ blast/Blast.cgi”. By sending different parameters to this API, the user can submit and retrieve queries that are executed by NCBI web servers. Every call to the API must include a CMD parameter that defines the type of the call. When submitting queries using ${\mathrm{CMD}}{=}\mathrm{Put}$ , the user can specify the querying database with the DATABASE parameter, the searching program with the PROGRAM parameter, and the query sequence with the QUERY parameter. The user will get an RID after the ${\mathrm{CMD}}{=}\mathrm{Put}$ API call, and can make another API call with the Get command and the returned RID to retrieve its BLAST results.
BLAST URL API。BLAST 接受核苷酸或氨基酸序列作为输入,并在数据库中查找最相似的序列 (Altschul et al., 1990; Boratyn et al., 2013)。结果可用于推断序列间关系或识别基因家族成员。该API允许用户通过NCBI服务器上的BLAST算法提交查询,以发现核苷酸或蛋白质序列与现有数据库之间的相似区域。BLAST URL API的基础地址为"https://blast.ncbi.nlm.nih.gov/blast/Blast.cgi"。通过向该API发送不同参数,用户可提交并获取由NCBI网络服务器执行的查询结果。每次API调用必须包含定义调用类型的CMD参数。使用 ${\mathrm{CMD}}{=}\mathrm{Put}$ 提交查询时,用户可通过DATABASE参数指定查询数据库,用PROGRAM参数选择搜索程序,用QUERY参数设置查询序列。用户将在 ${\mathrm{CMD}}{=}\mathrm{Put}$ 调用后获得RID,随后可通过Get命令和返回的RID发起另一API调用来获取BLAST结果。
2.2 In-context learning
2.2 上下文学习
We teach an LLM to use NCBI Web APIs through in-context learning with an engineered prompt. Figure 1 shows an example of the GeneGPT prompt, which is composed of four modules: 1. an instruction; 2. API documentation s; 3. API demonstra- tions; 4. a test question. The first three parts are fixed for all tasks, while the last one is task-specific.
我们通过上下文学习的方式,使用精心设计的提示词(prompt)教授大语言模型(LLM)调用NCBI网络API。图1: GeneGPT提示词示例,该提示词由四个模块组成:(1) 指令;(2) API文档;(3) API调用示例;(4) 测试问题。前三个部分对所有任务保持固定,最后一部分则根据具体任务进行调整。
- Instruction: The prompt starts with an overall task description (“Your task is to use NCBI APIs to answer genomic questions.”). It is then followed by documentation s and demonstrations of API usage summarized in Table 1.
- 指令:提示以整体任务描述开头("你的任务是使用NCBI API回答基因组学问题"),随后是文档说明和表1中总结的API使用示例。
- Documentation s (Dc.) provide natural language descriptions of the API functionality, general syntax, and argument choices. We include one for the E-utils API (Dc.1) and one for the BLAST tool (Dc.2).
- 文档 (Dc.) 提供API功能的自然语言描述、通用语法及参数选项。我们包含了一份E-utils API文档 (Dc.1) 和一份BLAST工具文档 (Dc.2)。
Table 1: Summary of API usage documentation s (Dc.1 and Dc.2) and demonstrations (Dm.1-4) in the GeneGPT prompt. Complete texts are shown in Appendix A.
| Comp. | Documentation | Database | Function |
| Dc.1 | E-utils | gene, | esearch, efetch, |
| snp, omim | esummary | ||
| Dc.2 | BLAST | nt | blastn |
| Comp. | Demonstration | Database | Function |
| Dm.1 | Alias | gene | esearch-> efetch |
| Dm.2 | Gene SNP | snp | esummary |
| Dm.3 | Gene disease | omim | esearch-> esummary |
| Dm.4 | Alignment | nt | blastn |
表 1: GeneGPT提示中API使用文档(Dc.1和Dc.2)及演示(Dm.1-4)的摘要。完整文本见附录A。
| 组件 | 文档 | 数据库 | 功能 |
|---|---|---|---|
| Dc.1 | E-utils | gene, | esearch, efetch, |
| snp, omim | esummary | ||
| Dc.2 | BLAST | nt | blastn |
| 组件 | 演示 | 数据库 | 功能 |
| Dm.1 | Alias | gene | esearch-> efetch |
| Dm.2 | Gene SNP | snp | esummary |
| Dm.3 | Gene disease | omim | esearch-> esummary |
| Dm.4 | Alignment | nt | blastn |
- Demonstrations (Dm.) are concrete examples of using NCBI Web APIs to solve ques- tions. Based on questions in the GeneTuring tasks, we manually write four demonstra- tions that cover four functions (esearch, efetch, esummary, blastn) and four databases (gene, snp, omim, nt) of E-utils and BLAST. The API URLs and the call results are marked up by “[ ]”, with a special “->” symbol inserted in between that serves as an indicator for API calls.
- 演示 (Dm.) 是使用NCBI Web API解决具体问题的实际案例。基于GeneTuring任务中的问题,我们手动编写了四个演示,涵盖E-utils和BLAST的四个功能 (esearch、efetch、esummary、blastn) 以及四个数据库 (gene、snp、omim、nt)。API URL和调用结果用“[ ]”标注,中间插入特殊的“->”符号作为API调用的指示符。
- Test question: The specific test question is then appended to the end of the prompt.
- 测试问题:将具体的测试问题附加到提示词末尾。
While the initial GeneGPT uses all documentations and demonstrations (denoted as GeneGPTfull in Table 2), we find through analyses in $\S4.1$ that GeneGPT can work well with only two demonstrations (denoted as GeneGPT-slim) on all tasks.
虽然最初的GeneGPT使用了所有文档和演示(在表2中记为GeneGPTfull),但我们通过$\S4.1$的分析发现,GeneGPT仅需两个演示(记为GeneGPT-slim)即可在所有任务上表现良好。
2.3 Inference algorithm
2.3 推理算法
The GeneGPT inference algorithm is briefly shown in Algorithm 1. Specifically, we first append the given question to the prompt (described in $\S2.2)$ and feed the concatenated text to Codex (code-davinci-002, Chen et al. (2021)) with a temperature of 0. We choose to use Codex for two reasons: (1) it is pre-trained with code data and shows better code understanding abilities, which
GeneGPT的推理算法如算法1所示。具体而言,我们首先将给定问题附加到提示词(详见$\S2.2$),然后将拼接后的文本输入温度为0的Codex(code-davinci-002,Chen等人(2021))。选择Codex基于两个原因:(1)其预训练包含代码数据,展现出更优的代码理解能力
Algorithm 1 GeneGPT inference algorithm
算法 1: GeneGPT 推理算法
| Input: question Model: Codex (code-davinci-002) Output: answer prompt ← header + demonstrations + question |
| finished ← False while not finished do next token ← Codex(prompt) |
| prompt ← prompt + next token if next token is "->" then |
| url ← extractLastURL(prompt) result ← callWebAPI(url) |
| prompt ← prompt + result |
| else if next token is "\n\n"then |
| answer extractAnswer(prompt) |
| finished ← True |
| end if |
| 输入: 问题 模型: Codex (code-davinci-002) 输出: 答案 |
| prompt ← 头部信息 + 示例 + 问题 |
| finished ← False while not finished do next token ← Codex(prompt) |
| prompt ← prompt + next token if next token is "->" then |
| url ← extractLastURL(prompt) result ← callWebAPI(url) |
| prompt ← prompt + result |
| else if next token is "\n\n"then |
| answer ← extractAnswer(prompt) |
| finished ← True |
| end if |
is crucial in generating the URLs and interpreting the raw API results; (2) its API has the longest (8k tokens) context length among all available models so that we can fit the demonstrations in.
在生成URL和解析原始API结果时至关重要;(2) 其API在所有可用模型中拥有最长的上下文长度(8k tokens),因此我们可以将演示内容完整放入。
We discontinue the text generation process when the special “ $\qquad\cdot>\quad,$ ” symbol is detected, which is the indication for an API call request. Then we extract the last URL and call the NCBI Web API with it. The raw execution results will be appended to the generated text, and it will be fed to Codex to continue the generation. When $\cdots(n\setminus\Gamma^{\prime\prime})$ , an answer indicator used in the demonstrations, is generated, we will stop the inference and extract the answer after the generated “Answer: ”.
当检测到特殊的 " $\qquad\cdot>\quad,$ " 符号时,我们会终止文本生成过程,该符号表示API调用请求。随后提取最后一个URL并用其调用NCBI Web API。原始执行结果会被追加到生成的文本中,并输入给Codex以继续生成。当生成演示中使用的答案指示符 $\cdots(n\setminus\Gamma^{\prime\prime})$ 时,我们将停止推理并提取"Answer: "后的答案。
3 Experiments
3 实验
3.1 GeneTuring
3.1 GeneTuring
The GeneTuring benchmark (Hou and Ji, 2023) contains 12 tasks, and each task has 50 questionanswer pairs. We use 9 GeneTuring tasks that are related to NCBI resources to evaluate the proposed GeneGPT model, and the QA samples are shown in Appendix B. The chosen tasks are classified into four modules and briefly described in this section.
GeneTuring基准测试 (Hou and Ji, 2023) 包含12项任务,每项任务有50组问答对。我们选用其中9个与NCBI资源相关的GeneTuring任务来评估提出的GeneGPT模型,问答样本见附录B。所选任务分为四个模块,本节将简要描述。
Nomenclature: This is about gene names. We use the gene alias task and the gene name conversion task, where the objective is to find the official gene symbols for their non-official synonyms.
术语说明:本文涉及基因命名。我们采用基因别名任务和基因名称转换任务,目标是为非官方同义词找到官方基因符号。
Genomics location: The tasks are about the locations of genes, single-nucleotide polymorphism (SNP), and their relations. We include the gene location, SNP location, and gene SNP association tasks. The first two tasks ask for the chromosome locations (e.g., “chr2”) of a gene or an SNP, and the last one asks for related genes for a given SNP.
基因组学定位:任务涉及基因、单核苷酸多态性(SNP)及其相互关系的位置。我们包含基因定位、SNP定位和基因-SNP关联任务。前两项任务要求提供基因或SNP的染色体位置(例如"chr2"),最后一项任务要求找出给定SNP的相关基因。
Functional analysis: It asks for gene functions. We use the gene disease association task where the goal is to return related genes for a given disease, and the protein-coding genes task which asks whether a gene is a protein-coding gene or not.
功能分析:该任务涉及基因功能研究。我们采用基因-疾病关联任务(目标是返回与给定疾病相关的基因)和蛋白质编码基因判断任务(判定某基因是否为蛋白质编码基因)。
Sequence alignment: The tasks query specific DNA sequences. We use the DNA sequence alignment to human genome task and the DNA sequence alignment to multiple species task. The former maps an DNA sequence to a specific human chromosome, while the latter maps an DNA sequence to a specific species (e.g. “zebrafish”).
序列比对:任务查询特定DNA序列。我们使用人类基因组DNA序列比对任务和多物种DNA序列比对任务。前者将DNA序列映射到特定人类染色体,后者将DNA序列映射到特定物种(如"斑马鱼")。
3.2 Compared methods
3.2 对比方法
We evaluate two settings of GeneGPT, a full setting (GeneGPT-full) where all prompt components are used, as well as a slim setting (GeneGPT-slim) inspired by our ablation and probing analyses (§4.1) where only Dm.1 and Dm.4 are used.
我们评估了GeneGPT的两种设置:完整设置(GeneGPT-full)使用所有提示组件,以及精简设置(GeneGPT-slim)仅使用Dm.1和Dm.4(灵感来自消融和探测分析(§4.1))。
We compare GeneGPT with various baselines evaluated by Hou and Ji (2023), including generaldomain GPT-based (Radford et al., 2018) LLMs such as GPT-2 (Radford et al., 2019), GPT-3 (text-davinci-003) (Brown et al., 2020), and ChatGPT5, GPT-2-sized biomedical domainspecific LLMs such as BioGPT (Luo et al., 2022) and BioMedLM6, as well as the new $\mathrm{Bing^{7}}$ , a retrieval-augmented LLM that has access to relevant web pages retrieved by Bing.
我们将GeneGPT与Hou和Ji (2023) 评估的多种基线进行比较,包括通用领域的基于GPT (Radford et al., 2018) 的大语言模型,如GPT-2 (Radford et al., 2019)、GPT-3 (text-davinci-003) (Brown et al., 2020) 和ChatGPT5,以及GPT-2规模的生物医学领域专用大语言模型,如BioGPT (Luo et al., 2022) 和BioMedLM6,还有新推出的$\mathrm{Bing^{7}}$,这是一种检索增强的大语言模型,可以访问由Bing检索到的相关网页。
3.3 Evaluation
3.3 评估
For the performance of the compared methods, we directly use the results reported in the original benchmark that are manually evaluated.
对于对比方法的性能表现,我们直接采用了原基准测试中人工评估报告的结果。
To evaluate our proposed GeneGPT method, we follow the general criteria but perform automatic evaluations. Specifically, we only consider exact matches between model predictions and the ground truth as correct predictions for all nomenclature and genomics location tasks. For the gene disease association task, we measure the recall as in the original dataset but based on exact individual gene matches. For the protein-coding genes task and the DNA sequence alignment to multiple species task, we also consider exact matches as correct after applying a simple vocabulary mapping that converts model-predicted “yes”/“no” to “TRUE”/“NA” and Latin species names to their informal names (e.g., “Saccharomyces cerevisiae” to “yeast”), respectively. For the DNA sequence alignment to human genome task, we give correct chromosome mapping but incorrect position mapping a score of 0.5 (e.g., chr8:7081648-7081782 v.s. chr8:1207812-1207946), since the original task does not specify a reference genome. Overall, our evaluation of GeneGPT is more strict than the original evaluation of other LLMs in Hou and Ji (2023), which performs manual evaluation and might consider non-exact matches as correct.
为评估我们提出的GeneGPT方法,我们遵循通用标准但采用自动评估方式。具体而言,对于所有命名法和基因组定位任务,仅当模型预测与标准答案完全匹配时才判定为正确预测。在基因疾病关联任务中,我们沿用原始数据集的召回率计算方式,但基于单个基因的精确匹配。对于蛋白质编码基因任务和DNA序列多物种比对任务,我们通过简单词汇映射(如将模型预测的"yes"/"no"转换为"TRUE"/"NA",将拉丁物种名转为常用名(例如"Saccharomyces cerevisiae"转为"yeast")后,同样采用精确匹配判定标准。在DNA序列人类基因组比对任务中,若染色体定位正确但位置错误(如chr8:7081648-7081782对比chr8:1207812-1207946),我们给予0.5分,因为原始任务未指定参考基因组。总体而言,我们的GeneGPT评估标准比Hou和Ji (2023)对其他大语言模型的人工评估更为严格,后者可能接受非精确匹配为正确答案。
| GeneTuring task | GPT-2 | BioGPT | BioMedLM | GPT-3 | ChatGPT | New Bing | GeneGPT (ours) | |
| -full | -slim | |||||||
| Nomenclature | ||||||||
| Gene alias | 0.00 | 0.00 | 0.04 | 0.09 | 0.07 | 0.66 | 0.80 * | 0.84 * |
| Gene name conversion | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.85 | 1.00 | 1.00 |
| Average | 0.00 | 0.00 | 0.02 | 0.05 | 0.04 | 0.76 | 0.90 | 0.92 |
| Genomic location | ||||||||
| Gene SNP association | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 * | 1.00 |
| Gene location | 0.01 | 0.04 | 0.12 | 0.09 | 0.09 | 0.61 | 0.62 | 0.66 |
| SNP location | 0.03 | 0.05 | 0.01 | 0.02 | 0.05 | 0.01 | 1.00 | 0.98 |
| Average | 0.01 | 0.03 | 0.04 | 0.04 | 0.05 | 0.21 | 0.87 | 0.88 |
| Functional analysis | ||||||||
| Gene disease association | 0.00 | 0.02 | 0.16 | 0.34 | 0.31 | 0.84 | 0.76 * | 0.66 |
| Protein-coding genes | 0.00 | 0.18 | 0.37 | 0.70 | 0.54 | 0.97 | 0.76 | 1.00 |
| Average | 0.00 | 0.10 | 0.27 | 0.52 | 0.43 | 0.91 | 0.76 | 0.84 |
| Sequence alignment | ||||||||
| DNA to human genome | 0.02 | 0.07 | 0.03 | 0.00 | 0.00 | 0.00 | 0.44 * | 0.44 * |
| DNA to multiple species | 0.02 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.86 | 0.88 |
| Average | 0.02 | 0.04 | 0.02 | 0.10 | 0.00 | 0.00 | 0.65 | 0.66 |
| Overall average | 0.00 | 0.04 | 0.08 | 0.16 | 0.12 | 0.44 | 0.80 | 0.83 |
Table 2: Performance of GeneGPT compared to other LLMs on the GeneTuring benchmark. \ * One-shot learning for GeneGPT. Bolded and underlined numbers denote the highest and second-highest performance, respectively.
| GeneTuring任务 | GPT-2 | BioGPT | BioMedLM | GPT-3 | ChatGPT | New Bing | GeneGPT (ours) | |
|---|---|---|---|---|---|---|---|---|
| -full | -slim | |||||||
| * * 命名规范 * * | ||||||||
| 基因别名 | 0.00 | 0.00 | 0.04 | 0.09 | 0.07 | 0.66 | 0.80 * | 0.84 * |
| 基因名称转换 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.85 | 1.00 | 1.00 |
| 平均 | 0.00 | 0.00 | 0.02 | 0.05 | 0.04 | 0.76 | 0.90 | 0.92 |
| * * 基因组定位 * * | ||||||||
| 基因SNP关联 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 * | 1.00 |
| 基因位置 | 0.01 | 0.04 | 0.12 | 0.09 | 0.09 | 0.61 | 0.62 | 0.66 |
| SNP位置 | 0.03 | 0.05 | 0.01 | 0.02 | 0.05 | 0.01 | 1.00 | 0.98 |
| 平均 | 0.01 | 0.03 | 0.04 | 0.04 | 0.05 | 0.21 | 0.87 | 0.88 |
| * * 功能分析 * * | ||||||||
| 基因疾病关联 | 0.00 | 0.02 | 0.16 | 0.34 | 0.31 | 0.84 | 0.76 * | 0.66 |
| 蛋白质编码基因 | 0.00 | 0.18 | 0.37 | 0.70 | 0.54 | 0.97 | 0.76 | 1.00 |
| 平均 | 0.00 | 0.10 | 0.27 | 0.52 | 0.43 | 0.91 | 0.76 | 0.84 |
| * * 序列比对 * * | ||||||||
| DNA到人类基因组 | 0.02 | 0.07 | 0.03 | 0.00 | 0.00 | 0.00 | 0.44 * | 0.44 * |
| DNA到多物种 | 0.02 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.86 | 0.88 |
| 平均 | 0.02 | 0.04 | 0.02 | 0.10 | 0.00 | 0.00 | 0.65 | 0.66 |
| * * 总体平均 * * | 0.00 | 0.04 | 0.08 | 0.16 | 0.12 | 0.44 | 0.80 | 0.83 |
表 2: GeneGPT与其他大语言模型在GeneTuring基准测试中的性能对比。 * GeneGPT采用单样本学习。加粗下划线数字分别表示最高和第二高表现。
3.4 Main results
3.4 主要结果
Table 2 shows the performance of GeneGPT on the GeneTuring tasks in comparison with other LLMs. For GeneGPT, tasks with “\ * ” in Table 2 are one-shot where one instance is used as API demonstration, and the other tasks are zero-shot. For the compared LLMs, all tasks are zero-shot.
表 2: GeneGPT 在 GeneTuring 任务上的性能与其他大语言模型的对比。对于 GeneGPT,表 2 中带“ * ”的任务是少样本 (one-shot) 任务,其中使用一个实例作为 API 演示,其他任务为零样本 (zero-shot) 任务。对于对比的大语言模型,所有任务均为零样本。
Nomenclature: GeneGPT achieves state-of-theart (SOTA) performance on both the one-shot gene alias task with an accuracy of 0.84 and the zeroshot gene name conversion task with an accuracy of 1.00. On average, GeneGPT outperforms New Bing by a large margin (0.92 v.s. 0.76). All other GPT models have accuracy scores of less than 0.10 on the nomenclature tasks.
术语说明:GeneGPT 在单次基因别名任务中准确率达到 0.84,零样本基因名称转换任务中准确率达到 1.00,均实现了最先进 (SOTA) 性能。GeneGPT 平均表现大幅优于 New Bing (0.92 对 0.76)。其他所有 GPT 模型在术语任务中的准确率均低于 0.10。
Genomic location: GeneGPT also achieves SOTA performance on all genomic location tasks, including the gene SNP association task (1.00) gene location task (0.66) and the SNP location task (1.00). While the New Bing is comparable to GeneGPT on gene location (0.61 v.s. 0.66), its performance on the two SNP-related tasks is close to 0. Again, most other LLMs score less than 0.10. Notably, while all genomics location tasks are zeroshot for GeneGPT-slim, it performs comparably to GeneGPT-full which uses one gene SNP association demonstration. This indicates that API demon- strations have strong cross-task general iz ability.
基因组位置:GeneGPT在所有基因组位置任务上也实现了SOTA性能,包括基因SNP关联任务(1.00)、基因位置任务(0.66)和SNP位置任务(1.00)。虽然New Bing在基因定位任务上与GeneGPT相当(0.61 vs. 0.66),但它在两个SNP相关任务上的表现接近于0。同样,大多数其他大语言模型得分低于0.10。值得注意的是,虽然所有基因组位置任务对GeneGPT-slim都是零样本学习,但其表现与使用一个基因SNP关联演示的GeneGPT-full相当。这表明API演示具有强大的跨任务泛化能力。
Functional analysis: The new Bing performs better functional analysis tasks than the proposed GeneGPT (average score: 0.91 v.s. 0.84), which is probably because many web pages related to gene functions can be retrieved by the Bing search engine. We also note that other LLMs, especially GPT-3 and ChatGPT, perform moderately well and much better than they perform on other tasks. This might also be due to the fact that many gene-function-related texts are included in their pre-training corpora.
功能分析:新版Bing在功能分析任务上表现优于提出的GeneGPT(平均得分:0.91 vs. 0.84),这可能是因为Bing搜索引擎能检索到许多与基因功能相关的网页。我们还注意到其他大语言模型,尤其是GPT-3和ChatGPT,表现中等且明显优于它们在其他任务上的表现。这也可能是由于它们的预训练语料中包含了许多与基因功能相关的文本。
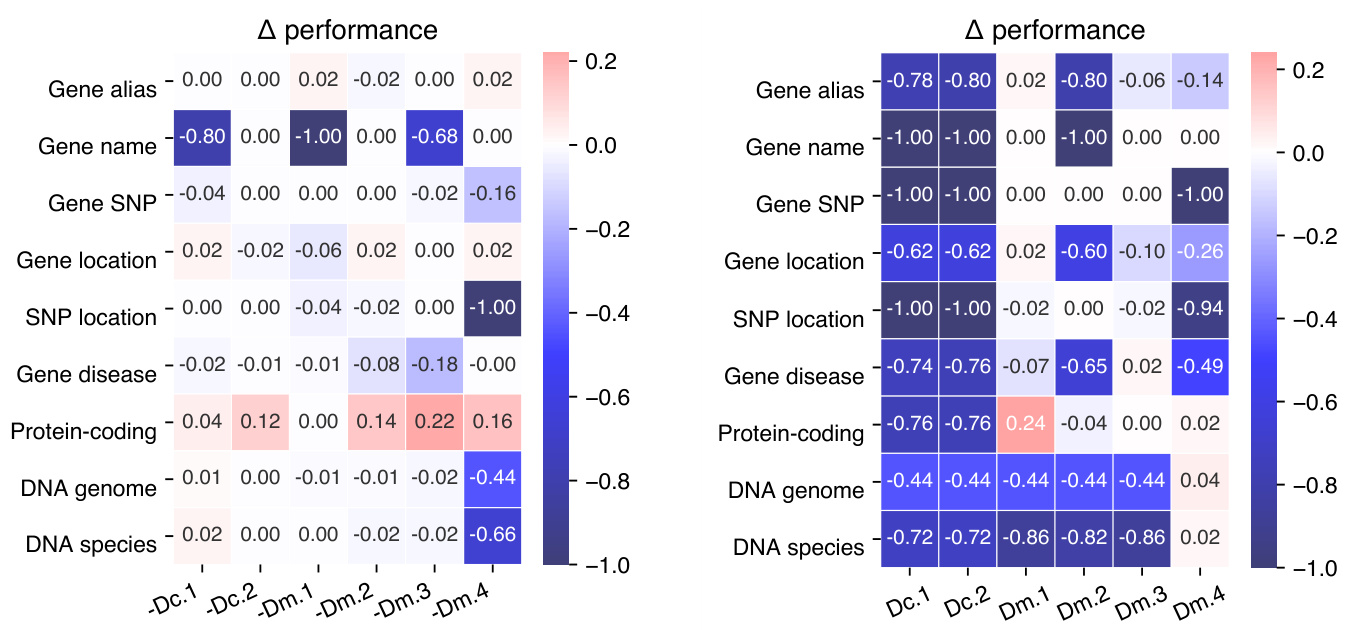
Figure 2: Performance changes of the ablation (left) and probing (right) experiments as compared to GeneGPTfull.
图 2: 消融实验(左)与探针实验(右)相对于GeneGPTfull的性能变化
Sequence alignment: GeneGPT performs much better with an average score of 0.66 than all other models including the new Bing (0.00), which essentially fails on the sequence alignment tasks. This is not very surprising since sequence alignment is easy with the BLAST tool, but almost impossible for an auto-regressive LLM even with retrieval augmentation as the input sequences are too specific to be indexed by a search engine.
序列比对:GeneGPT以0.66的平均得分显著优于包括新必应(0.00)在内的所有其他模型,后者在序列比对任务上基本失效。这一结果并不意外,因为使用BLAST工具进行序列比对很容易,但对于自回归大语言模型来说几乎不可能实现——即使采用检索增强技术,输入序列的特异性也使其难以被搜索引擎索引。
Although evaluated under a more strict setting (§3.3), GeneGPT achieves a macro-average performance of 0.83 which is much higher than other compared LLMs including New Bing (0.44). Overall, GeneGPT achieves new SOTA performance on all 2 one-shot tasks and 6 out of 7 zero-shot tasks and is outperformed by New Bing only on the gene disease association task.
尽管在更严格的设置下进行评估(§3.3),GeneGPT的宏观平均性能达到0.83,远高于包括New Bing(0.44)在内的其他对比大语言模型。总体而言,GeneGPT在所有2项单样本任务和7项零样本任务中的6项上实现了新的SOTA性能,仅在基因疾病关联任务上表现略逊于New Bing。
4.1 RQ1: Component importance
4.1 RQ1: 组件重要性
We conduct ablation and probing experiments to study the importance of individual prompt components, including 2 documentation s (Dc.1, Dc.2) and 4 demonstrations (Dm.1-4) described in $\S2.2$ .
我们通过消融和探查实验来研究各个提示组件的重要性,包括2个文档 (Dc.1, Dc.2) 和4个演示 (Dm.1-4) ,如 $\S2.2$ 所述。
For ablation tests, we remove each component from GeneGPT-full and then evaluate the prompt. The results are shown in Figure 2 (left). Notably, the performance on the DNA to genome and species alignment tasks is only significantly decreased without the BLAST demonstration (Dm.4), but not affected by the ablation of the BLAST documentation (Dc.2). While the ablations of other components decrease the performance, most only affect one relevant task (e.g., Dm.1 and gene name conversion), which indicates a high level of redundancy of the prompt components.
为了进行消融测试,我们从GeneGPT-full中逐一移除每个组件,然后评估提示效果。结果如图2 (左) 所示。值得注意的是,DNA到基因组和物种比对任务的性能仅在移除BLAST演示 (Dm.4) 时显著下降,但不受BLAST文档移除 (Dc.2) 的影响。虽然其他组件的移除会降低性能,但大多数只影响一个相关任务 (例如Dm.1和基因名称转换),这表明提示组件具有高度的冗余性。
4 Discussions
4 讨论
We have shown that GeneGPT largely surpasses various LLMs on the GeneTuring benchmark. In this section, we further characterize GeneGPT by studying three research questions (RQ):
我们已证明GeneGPT在GeneTuring基准测试中大幅超越多种大语言模型。本节通过研究三个研究问题(RQ)进一步刻画GeneGPT的特性:
RQ1: What is the importance of each prompt component in GeneGPT? RQ2: Can GeneGPT answer multi-hop questions by chain-of-thought API calls? RQ3: What types of errors does GeneGPT make on each studied task?
RQ1: GeneGPT中各提示组件的重要性是什么?
RQ2: GeneGPT能否通过思维链(chain-of-thought)API调用回答多跳问题?
RQ3: GeneGPT在每项研究任务中会犯哪些类型的错误?
For the probing experiments, we evaluate GeneGPT with only one prompt component to study the individual capability. The results are shown in Figure 2 (right). Overall, GeneGPT with only one documentation (Dc.1 or Dc.2) fails on all tasks. Surprisingly, with only one demonstration of the gene alias task (Dm.1) in the prompt, GeneGPT is able to perform comparably to GeneGPT-full on all tasks except the alignment ones. On the other hand, GeneGPT with only the BLAST demonstration (Dm.4) performs well on the two alignment tasks, which is somehow expected. These results suggest that GeneGPT with only two demonstrations (Dm.1 and Dm.4) in the prompt can generalize to all tasks in the GeneTuring benchmark. We denote this as GeneGPT-slim, and results in Table 2 show that with only two demonstrations, it outperforms the GeneGPT-full and achieves stateof-the-art overall results on GeneTuring.
在探测实验中,我们仅使用一个提示组件评估GeneGPT以研究其独立能力。结果如图2(右)所示。总体而言,仅包含一个文档(Dc.1或Dc.2)的GeneGPT在所有任务上均失败。令人惊讶的是,仅需在提示中包含一个基因别名任务示例(Dm.1),GeneGPT就能在除序列比对外的所有任务上达到与GeneGPT-full相当的表现。另一方面,仅包含BLAST示例(Dm.4)的GeneGPT在两个比对任务中表现良好,这符合预期。这些结果表明,提示中仅需两个示例(Dm.1和Dm.4)的GeneGPT即可泛化至GeneTuring基准的所有任务。我们将此版本记为GeneGPT-slim,表2结果显示仅需两个示例时,其性能超越GeneGPT-full并在GeneTuring上达到当前最佳总体结果。
4.2 RQ2: Multi-hop QA on GeneHop
4.2 RQ2: GeneHop上的多跳问答
Questions in the GeneTuring benchmark are singlehop and just require one step of reasoning, e.g., “Which gene is SNP rs 983419152 associated with?”. However, many real-world biomedical questions are multi-hop that need more steps to answer (Jin et al., 2022). For example, to answer “What is the function of the gene associated with SNP rs 983419152?”, the model should first get the associated gene name and then find its functions.
GeneTuring基准测试中的问题都是单跳的,仅需一步推理即可解答,例如"SNP rs983419152与哪个基因相关?"。然而现实中的许多生物医学问题属于多跳推理 (Jin et al., 2022),例如要回答"与SNP rs983419152相关基因的功能是什么?",模型需要先获取关联基因名称,再查询其功能。
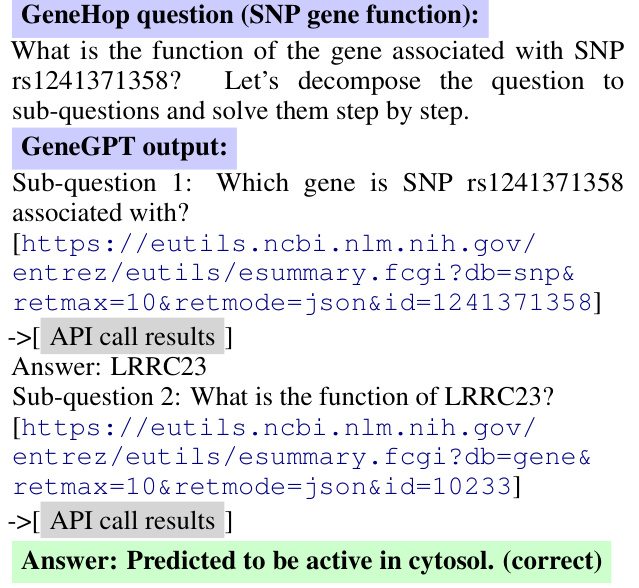
Figure 3: GeneGPT uses chain-of-thought API calls to answer a multi-hop question in GeneHop. Figure 4: GeneGPT uses chain-of-thought API calls to answer a multi-hop question in GeneHop.
图 3: GeneGPT 使用思维链 (chain-of-thought) API 调用来回答 GeneHop 中的多跳问题。
图 4: GeneGPT 使用思维链 (chain-of-thought) API 调用来回答 GeneHop 中的多跳问题。
To test GeneGPT’s capability of answering multi-hop questions, we present GeneHop, a novel dataset that contains three new multi-hop QA tasks based on the GeneTuring benchmark: (a) SNP gene function, which asks for the function of the gene associated with a given SNP. (b) Disease gene location, where the task is to list the chromosome locations of the genes associated with a given disease. (c) Sequence gene alias, which asks for the aliases of the gene that contains a specific DNA sequence. Each task in GeneHop contains 50 questions, and the collection pipeline is detailed in Appendix C. For all tasks, we append the chain-ofthought instruction “Let’s decompose the question to sub-questions and solve them step by step.” after the test question (Wei et al., 2022b).
为测试GeneGPT回答多跳问题的能力,我们提出了GeneHop数据集。该数据集基于GeneTuring基准包含三项新颖的多跳问答任务:(a) SNP基因功能,要求回答与给定SNP关联基因的功能;(b) 疾病基因定位,任务是列出与特定疾病相关基因的染色体位置;(c) 序列基因别名,要求找出包含特定DNA序列基因的别名。GeneHop中每个任务包含50个问题,具体收集流程详见附录C。所有测试问题后均附加思维链指令"让我们将问题分解为子问题并逐步解决" (Wei et al., 2022b)。
Figure 3 shows an example of GeneGPT to answer Task (a). In this case, GeneGPT successfully decomposes the multi-hop question into two sub-questions, and the sub-question 2 is based on the answer of the sub-question 1. Interestingly, GeneGPT uses a shortcut to answer sub-question 2: instead of first calling esearch and then calling esummary, GeneGPT finds the gene id in the API call results of sub-question 1 and directly calls esummary. This capability is not shown in the prompt but elicited by chain-of-thought API calls.
图 3 展示了 GeneGPT 回答任务 (a) 的示例。在该案例中,GeneGPT 成功将多跳问题分解为两个子问题,且子问题 2 的解答基于子问题 1 的答案。值得注意的是,GeneGPT 采用捷径回答子问题 2:它没有先调用 esearch 再调用 esummary,而是从子问题 1 的 API 调用结果中直接获取基因 ID 并调用 esummary。这种能力并未在提示中显式指定,而是通过思维链 (chain-of-thought) 式 API 调用所激发。
GeneHop question (Disease gene location):
基因定位问题(疾病基因位置):
List chromosome locations of the genes related to Cleft palate with an ky log loss i a. Let’s decompose the question to sub-questions and solve them step by step.
列出与腭裂相关基因的染色体位置及其ky对数损失值。让我们将问题分解为子问题并逐步解决。
GeneGPT output:
GeneGPT输出:
Figure 4 shows another example of GeneGPT answering Task (b), where GeneGPT successfully decomposes the multi-hop question and correctly calls the required APIs. Notably, the answering chain involves 3 sub-questions and 4 API calls, which are longer than all in-context demonstrations (1 single-hop question and 2 API calls at most). This ability to generalize to longer chains of thought is an important aspect of GeneGPT’s flexibility and usefulness for real-world applications.
图 4: GeneGPT 回答任务 (b) 的另一个示例,其中 GeneGPT 成功分解了多跳问题并正确调用了所需 API。值得注意的是,该回答链涉及 3 个子问题和 4 次 API 调用,比所有上下文演示 (最多 1 个单跳问题和 2 次 API 调用) 都要长。这种对更长思维链的泛化能力是 GeneGPT 在实际应用中灵活性和实用性的重要体现。
Table 3: Performance of multi-hop QA on GeneHop. We only compare GeneGPT with New Bing since other LLMs cannot even answer single-hop questions well.
| GeneHopTask | New Bing | GeneGPT |
| SNP genefunction | 0.00 | 0.55 |
| Disease gene location | 0.71 | 0.67 |
| Sequence gene alias | 0.00 | 0.28 |
| Average | 0.24 | 0.50 |
表 3: GeneHop多跳问答性能对比。由于其他大语言模型甚至无法较好回答单跳问题,我们仅比较GeneGPT与New Bing的表现。
| GeneHop任务 | New Bing | GeneGPT |
|---|---|---|
| SNP基因功能 | 0.00 | 0.55 |
| 疾病基因定位 | 0.71 | 0.67 |
| 序列基因别名 | 0.00 | 0.28 |
| 平均 | 0.24 | 0.50 |
We manually evaluate the results predicted by GeneGPT and compare it to the new Bing, which is the only baseline LLM that performs well on the single-hop GeneTuring benchmark due to its retrieval augmentation feature. The evaluation criteria are described in Appendix D. As shown in Table 3, while the new Bing outperforms GeneGPT on the disease gene location task, it is mostly using webpages that contain both the disease and location information without multi-hop reasoning. The new Bing fails to perform the other 2 tasks since the input information (SNP or sequence) is not indexed by Bing and can only be found in specialized databases. GeneGPT, on the other hand, performs moderately well on all 3 tasks, and achieves a much higher average score (0.50 v.s. 0.24).
我们手动评估了GeneGPT的预测结果,并将其与新版Bing进行对比。由于具备检索增强功能,新版Bing是唯一在单跳GeneTuring基准测试中表现优异的基础大语言模型。评估标准详见附录D。如表3所示,尽管新版Bing在疾病基因定位任务上优于GeneGPT,但其主要依赖同时包含疾病和位置信息的网页,并未进行多跳推理。由于输入信息(SNP或序列)未被Bing索引且仅存在于专业数据库中,新版Bing无法完成另外两项任务。相比之下,GeneGPT在所有三项任务中均表现稳定,并获得了显著更高的平均分(0.50对比0.24)。
4.3 RQ3: Error analysis
4.3 RQ3: 错误分析
We manually study all errors made by GeneGPT and classify them into five types. Table 4 shows the count of each error type on the evaluate tasks: E1: using the wrong API or not using APIs, e.g., using the gene instead of the omin database for diseases; E2: using the right API but wrong arguments, e.g., passing terms to id; E3: not extracting the answer in the API result, most commonly seen in gene function extraction; E4: right API call but results do not contain the answer, where the question is not answerable with NCBI databases; and O includes other unclassified errors. Specific error examples are shown in Appendix E.
我们人工研究了GeneGPT的所有错误,并将其分为五类。表4展示了各项评估任务中每种错误类型的数量:E1:使用错误的API或未使用API,例如针对疾病使用gene而非omin数据库;E2:使用正确API但参数错误,例如向id传递terms;E3:未从API结果中提取答案,最常见于基因功能提取任务;E4:API调用正确但结果不包含答案,即NCBI数据库无法回答该问题;O代表其他未分类错误。具体错误示例见附录E。
Our results suggest that different tasks have specific and enriched error types: simple tasks (alias and location) fail mostly because of E4; E1 only happens in disease-related tasks; alignment tasks face more issues with BLAST interfaces and reference genomes (O); multi-hop tasks in GeneHop tend to have E2 and E3 in the reasoning chains.
我们的结果表明,不同任务具有特定且丰富的错误类型:简单任务(别名和位置)失败主要由于E4;E1仅发生在疾病相关任务中;比对任务更多面临BLAST接口和参考基因组(O)问题;GeneHop中的多跳任务在推理链中容易出现E2和E3。
5 Related work
5 相关工作
Large language models: Recent studies have shown that scaling pre-trained LMs leads to performance improvement and potentially emergent abilities on various NLP tasks (Brown et al., 2020; Kaplan et al., 2020; Wei et al., 2022a; Chowdh- ery et al., 2022; OpenAI, 2023). However, such auto-regressive LLMs are still susceptible to hallucinations and generate erroneous content (Ji et al., 2023). Augmenting LLMs with external tools is a possible solution to this issue (Mialon et al., 2023).
大语言模型:近期研究表明,扩大预训练语言模型(LM)的规模能提升各类自然语言处理(NLP)任务的性能,并可能激发涌现能力 (Brown et al., 2020; Kaplan et al., 2020; Wei et al., 2022a; Chowdh- ery et al., 2022; OpenAI, 2023)。但这类自回归大语言模型仍易产生幻觉并生成错误内容 (Ji et al., 2023),通过外部工具增强大语言模型是潜在解决方案之一 (Mialon et al., 2023)。
Tool augmentation: Potential tools include: (1) search engines (Guu et al., 2020; Lewis et al., 2020; Borgeaud et al., 2022), also known as retrieval augmentation, exemplified by New Bing; (2) program APIs by in-context learning (Gao et al., 2022; Schick et al., 2023) or fine-tuning (Parisi et al., 2022; Schick et al., 2023). We present the first study on the in-context learning abilities of documentations and demonstrations of NCBI Web APIs.
工具增强:潜在工具包括:(1) 搜索引擎 (Guu et al., 2020; Lewis et al., 2020; Borgeaud et al., 2022),也称为检索增强,以New Bing为例;(2) 通过上下文学习 (Gao et al., 2022; Schick et al., 2023) 或微调 (Parisi et al., 2022; Schick et al., 2023) 实现的程序API。我们首次研究了NCBI Web API文档和演示的上下文学习能力。
Biomedical question answering: It is an essential step in clinical decision support (Ely et al., 2005) and biomedical knowledge acquisition (Jin et al., 2022). LLMs have been successfully applied to various biomedical QA tasks that are knowledgeor reasoning-intensive (Singhal et al., 2022; Liévin et al., 2022; Nori et al., 2023). However, autoregressive LLMs fail to perform data-intensive tasks which require the model to precisely store and recite database entries, such as the GeneTuring benchmark (Hou and Ji, 2023). Retrieval augmentation also falls short since specialized databases are usually not indexed by commercial search engines. GeneGPT solves this task by tool augmentation.
生物医学问答:这是临床决策支持 (Ely et al., 2005) 和生物医学知识获取 (Jin et al., 2022) 的关键环节。大语言模型已成功应用于多种知识密集或推理密集的生物医学问答任务 (Singhal et al., 2022; Liévin et al., 2022; Nori et al., 2023)。然而,自回归大语言模型难以胜任需要精确存储和复述数据库条目的数据密集型任务,例如GeneTuring基准测试 (Hou and Ji, 2023)。检索增强方法同样存在局限,因为专业数据库通常未被商业搜索引擎收录。GeneGPT通过工具增强解决了这一难题。
| GeneTuring Task | E1 | E2 | E3 | E4 | 0 |
| Gene alias | 0 | 0 | 2 | 6 | 0 |
| Genelocation | 0 | 0 | 0 | 17 | 0 |
| SNPlocation | 0 | 1 | 0 | 0 | 0 |
| Genediseaseassociation | 15 | 0 | 0 | 3 | 2 |
| DNA tohumangenome | 0 | 0 | 7 | 0 |
